Extended Data Figure 5: The training sample size needed to reach a great predictive performance scales linearly with the number of species in synthetic data for MelonnPan, MiMeNet, and mNODE.
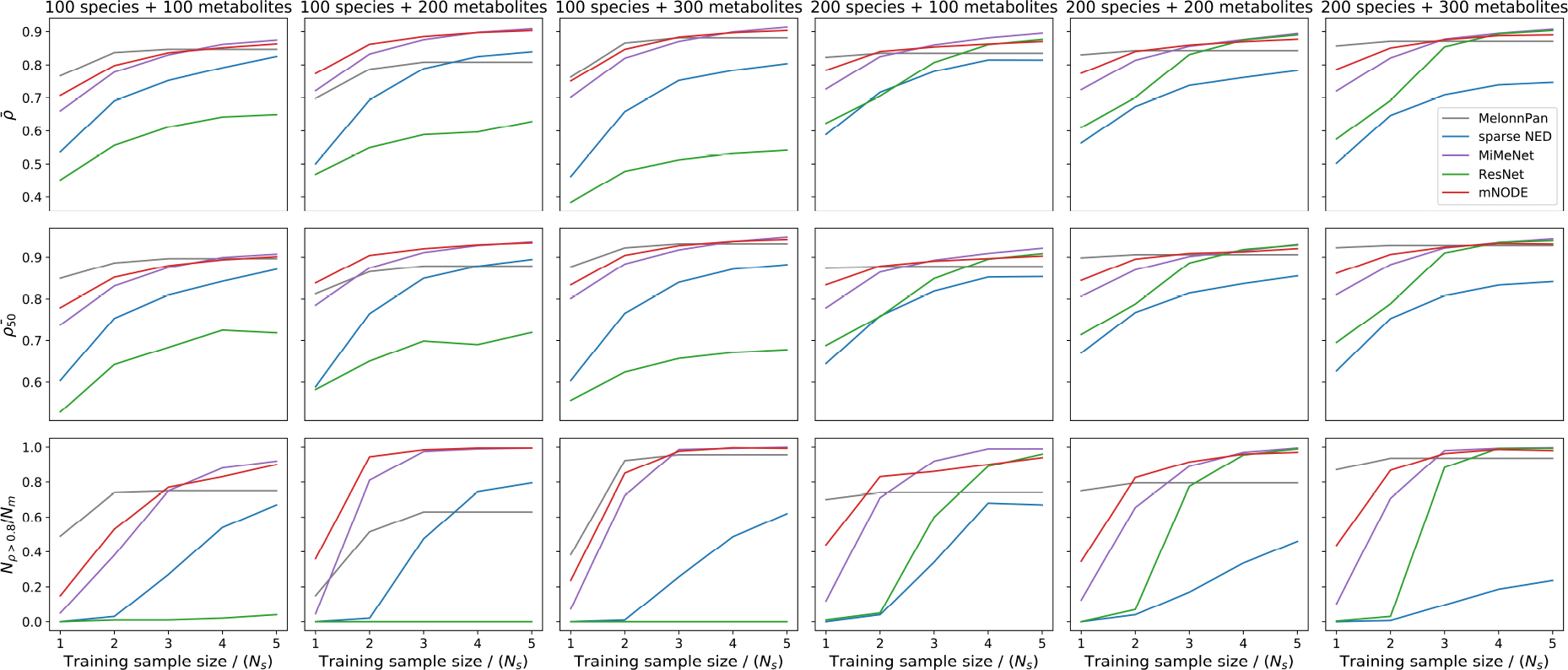
Synthetic data in this figure are generated by the microbial consumer-resource model with nutrient sampling probability . For the case with 100 species and varying number of metabolites (100, 200, or 300), three metrics are used for comparing model performances: the mean SCC , the top-50 mean SCC , and the number of metabolites with SCCs larger than 0.8 divided by the number of metabolites .
