Figure 3. Substrate recognition mechanisms of HDAC complexes.
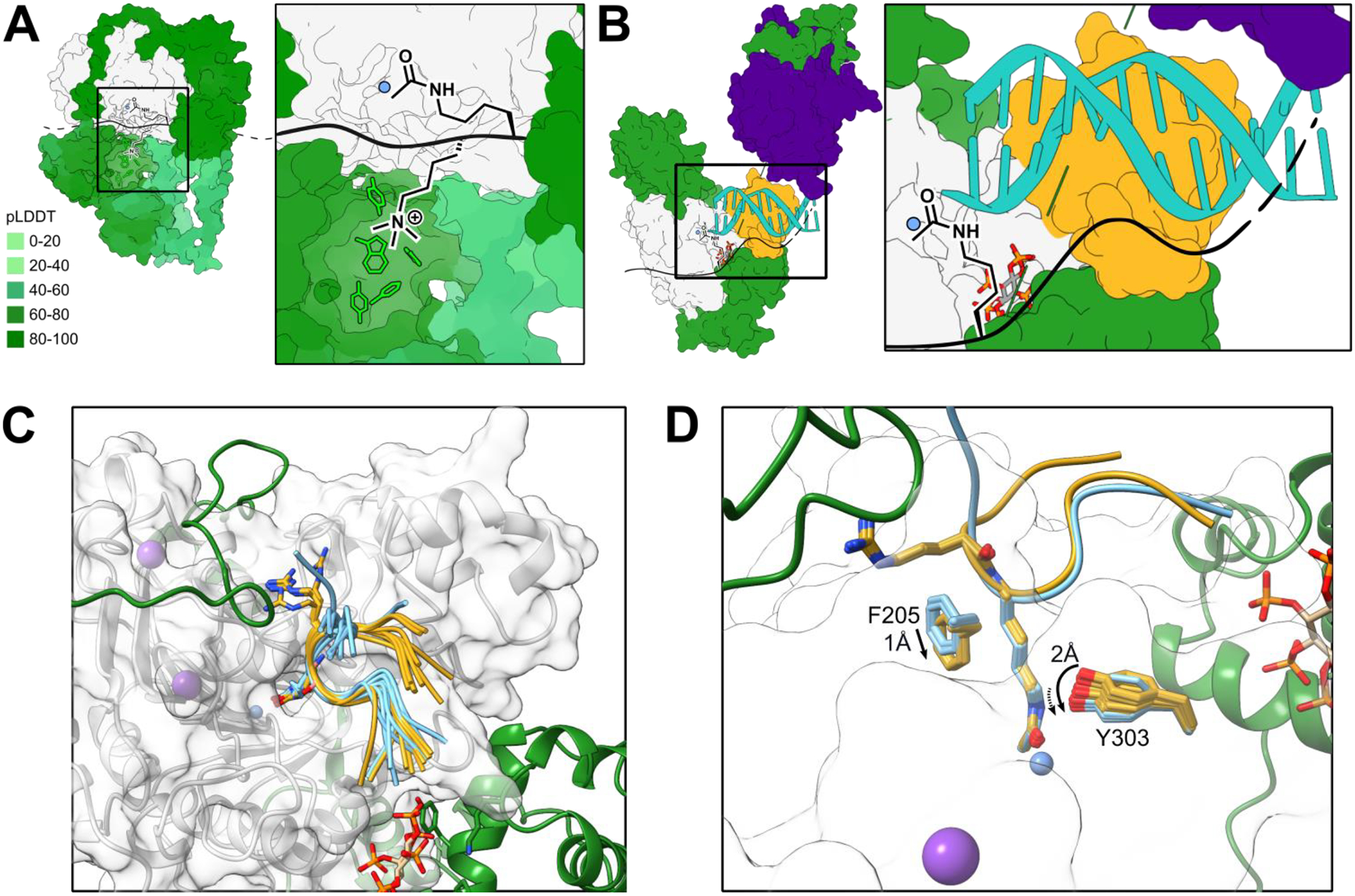
(A) RERE-HDAC1 (green-grey) complex with IP6 (red/orange/dark grey/white), potassium (purple) and zinc (blue). Inset: the methyllysine-binding aromatic cage (lime) of the BAH domain is depicted in complex with a theoretical methyllysine/acetyllysine substrate. Complex simulated with AlphaFold multimer (v 2.1.1); pLDDT quintiles depicted by green gradient with darker color corresponding to higher pLDDT. Top-scoring models were selected and potassium, zinc, and inositol hexaphosphate were added based on an existing model (PDBID: 5ICN), then subjected to Rosetta relax for further refinement. The lowest energy structure was selected for depiction. (B) Partial NuRD deacetylase module shown here with 1 each of MTA1-HDAC1-MBD2-RBBP4 (green-grey-gold-indigo; PDBID: 7AOA) complex with IP6 (red/orange/dark grey/white), potassium (purple) and zinc (blue), aligned MBD2-DNA complex (turquoise; PDBID: 7MWM). Inset: nucleic acid binding by the MBD domain could serve to position the HDAC active site near histone tails or other chromatin-bound proteins. (C-D) Histone H3 tail (gold and blue) docking to RCOR1elm2sant-HDAC1 (green-grey) complex illustrating conformational states dependent on the amino acid preceding the substrate lysine. Histone H3 peptide (S10-K18) was re-constructed from the existing peptide inhibitor-bound HDAC1 crystal structure (PDB 5ICN) using ChimeraX and Rosetta. Then, the reconstructed peptide was docked into the RCOR1elm2sant-HDAC1 complex structure generated by Alphafold multimer using Rosetta Flexpepdock [34]. Top 10 structures were superimposed for visual inspection (C). The lowest energy complex structure was further refined using Rosetta relax [31]. Sidechains of Phe205 and Tyr303 in the active site from all 50 relaxed structures were superimposed (D) to inspect conformational heterogeneity caused by G13R. (C) H3 tail sequences with either G13 preceding K14ac (blue) or R13 preceding K14ac (yellow) are shown. The conformational space surveyed by the arginine side chain contributes to a difference in the conformation of both HDAC and the C-terminal portion of the docked peptide. (D) Differences in active site residue positions and conformational heterogeneity dependent on the amino acid preceding the substrate lysine. Substitution of H3G13 for R results in a shift in the F205, a gatekeeper of the HDAC active site, and a change in dynamics of Y303, a hydrogen bond donor in catalysis. (A-D) All structures rendered with ChimeraX.
