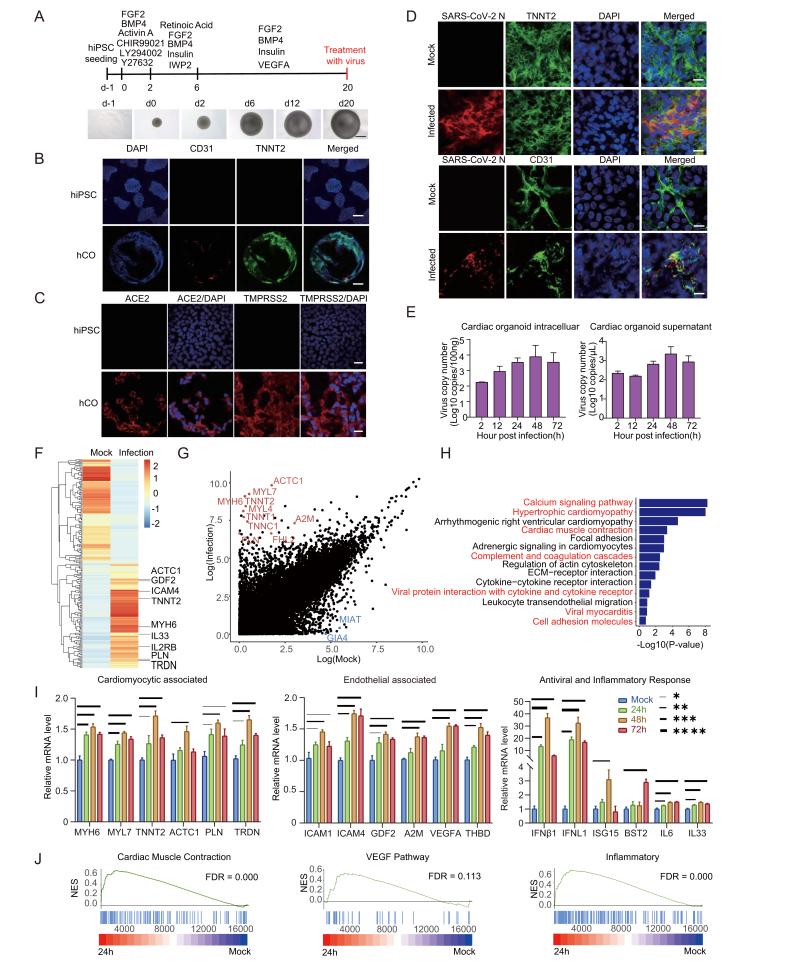
Fig. 1.
Overview of experimental protocol and results of 3D cardiac organoids culture. A Cardiac organoids were generated by step-wise differentiation of human-induced pluripotent stem cells. Scale bar: 200 μm. B Cell composition and distribution of cardiac organoids were assessed using immunofluorescence whole-mount microscopy. Scale bar: 100 μm. C Immunofluorescence staining for ACE2 and TMPRSS2 expression in hCOs and hiPSCs. Scale bar: 20 μm. D Immunofluorescence staining for SARS-CoV-2 N protein and cardiomyocyte marker TNNT2, as well as SARS-CoV-2 N protein and endothelial cell marker CD31. The hCOs were fixed by paraformaldehyde at 2 days post infection. Scale bar: 10 μm. E Viral RNA copies were quantified in SARS-CoV-2 infected cardiac organoids and the supernatant. F Heatmap showing differentially expressed genes (DEGs) in SARS-CoV-2-infected organoids (24 h) versus mock. G Differential normalized log2 expression in SARS-CoV-2-infected organoids (24 h) versus the mock infected organoids. H KEGG analysis of DEGs in SARS-CoV-2-infected organoids (24 h) versus mock infected organoids. I the cardiac organoids after SARS-CoV-2 infection were harvested to examine the expression of indicated genes using qRT-PCR. GAPDH was used as an internal control. Data were presented as mean ± SD. ∗ indicates P < 0.05; ∗∗ indicates P < 0.01; ∗∗∗ indicates P < 0.001. ∗∗∗∗ indicates P < 0.0001. J GSEA enrichment analysis of SARS-CoV-2-infected organoids (24 h) versus mock-infected organoids. Please see the Supplementary data Section for full experimental details.