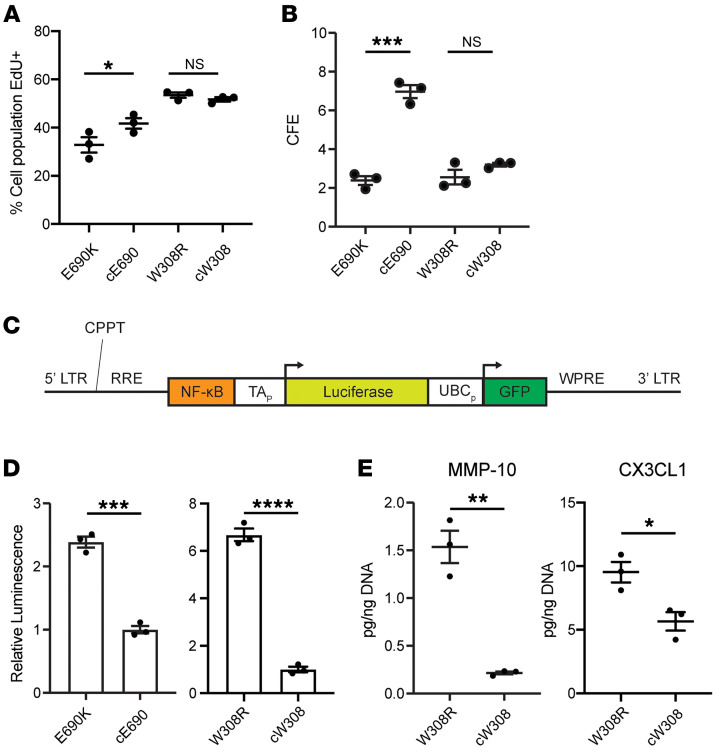
Figure 6. iAEC2s expressing E690K and W308R ABCA3 mutant proteins have increased canonical NFκB signaling activity and variable proliferative capacity.
(A) Percentages of day 75 patient iAEC2s incorporating EdU after 24 hour incubation. Biological replicates n = 3. *P ≤ 0.05, 2-tailed Student’s t test. (B) CFE reported as percentages of total spheres divided by input cells, of indicated patient iAEC2s on day 57. Biological replicates (n = 3), separated at day 0. Bars represent mean ± SE. ***P ≤ 0.001, ns, not significant, 2-tailed Student’s t test. (C) Lentiviral vector containing 2 gene expression cassettes, one allowing for constitutive expression of GFP for purifying transduced cells, and the second expressing luciferase under the regulatory control of a minimal promoter (TAp) adjacent to p50/p65 heterodimer consensus binding sequence (NFκB). This vector (detailed in Alysandratos et al., 2021 [36]) was used to transduce each indicated iAEC2 sample. LTR, lentiviral long terminal repeat; RRE, Rev responsive element; CPPT, central polypurine tract; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element; UBCp, ubiquitin protein-C promoter. (D) Relative NFκB pathway activity of lentiviral-transduced (GFP sorted) day 286 W308R and cW308 iAEC2s and day 158 E690K and cE690 iAEC2s measured by levels of D-luciferin luminescence. Replicates (n = 3), W308R and cW308 separated at day 148, E690K and cE690 separated at day 0. Graphs show mean ± SE. ***P ≤ 0.001, ****P ≤ 0.0001, 2-tailed Student’s t test. (E) Levels of indicated cytokines released in the culture supernatants of 2D monolayer cultured patient iAEC2s. Biological replicates (n = 3), separated at day 0. Graphs show mean ± SE. *P ≤ 0.05, **P ≤ 0.01, 2-tailed Student’s t test.