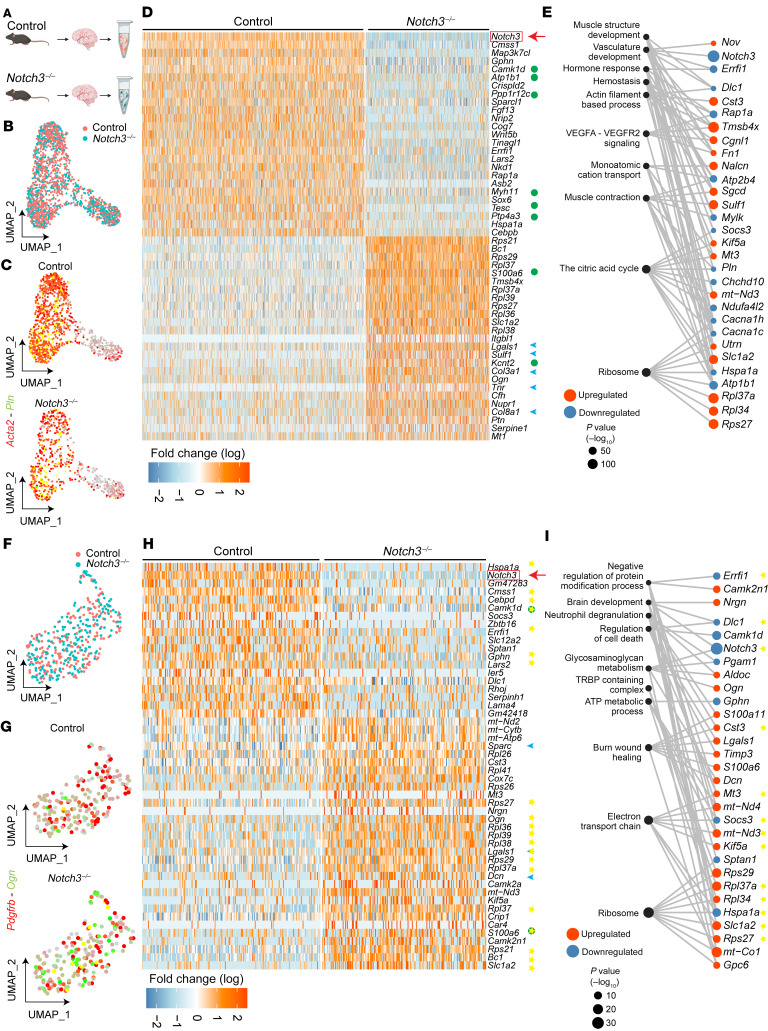
Figure 2. Loss of Notch3 in VSMCs and pericytes results in a decrease in transcripts that regulate contractility and an increase in transcripts associated with extracellular matrix.
(A) Schema of experimental design. Superficial brain vessels and penetrating brain arteries were dissected from mature (12-month-old) female and male Notch3–/– and littermate control mice and enzymatically dissociated to obtain single-cell suspensions for scRNA-Seq. n = 8 total mice, 2 female and 2 male pooled per genotype. (B) Uniform manifold approximation and projection (UMAP) plot of scRNA-Seq visualizing spread of data from VSMCs of the 2 genotypes. (C) Feature plot identifies Acta2+Pln+ VSMCs in control and Notch3–/–. (D) Heatmap visualizing the top 50 DEGs identified in VSMCs. (E) Gene Ontology enrichment of Notch3–/– VSMCs from the top 10 unique ontology categories and selected member genes within each category. Dot color indicates direction of expression change upon loss of Notch3–/–, while size indicates significance of enrichment. (F) UMAP plot of scRNA-Seq visualizing spread of data from pericytes of the 2 genotypes at 12 months of age. (G) Feature plot identifies Pdgfrb+Ogn+ pericytes in control and Notch3–/–. (H) Heatmap visualizing the top 50 DEGs identified in pericytes. (I) Gene Ontology enrichment of Notch3–/– pericytes from the top 10 unique ontology categories and selected member genes within each category. Dot color indicates direction of expression change upon loss of Notch3–/–, while size indicates significance of enrichment. Yellow stars indicate genes identified as DEGs in the same direction in both Notch3–/– VSMCs and Notch3–/– pericytes. For D and H, green circles indicate genes that regulate muscle cell contraction; blue arrowheads indicate ECM transcripts. Yellow stars indicate genes identified as DEGs in the same direction in both Notch3–/– VSMCs and Notch3–/– pericytes.