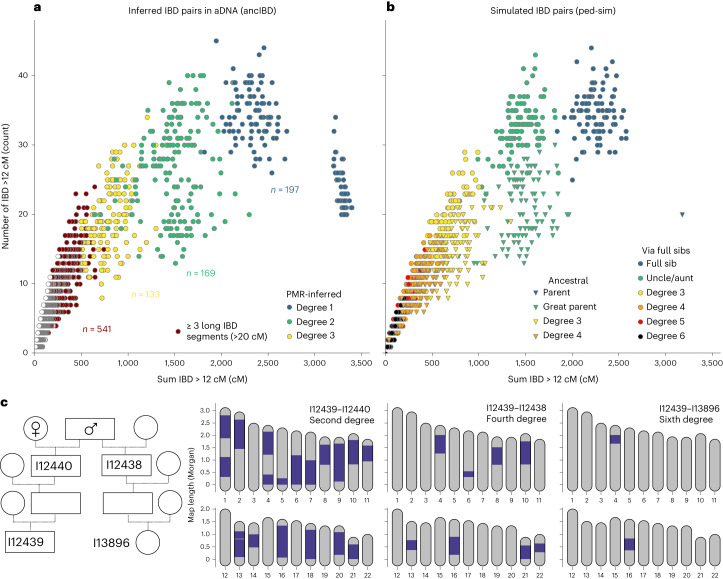
Fig. 3. Inferring biological relatives in the aDNA record using long IBD inferred with ancIBD.
a, Inferred IBD among pairs of 4,248 ancient Eurasian individuals. The plot visualizes both the count (y axis) as well as the summed length (x axis) of all IBD >12 cM long. For comparison, we colour-code pairs on the basis of relatedness estimates from pairwise mismatch rates (PMR) that can detect up to third-degree relatives (Supplementary Note 9). We also annotate new relatives found by ancIBD, indicated by at least three very long IBD segments (>20 cM) typical of up to sixth-degree relatives. b, Simulated IBD among pairs of relatives. For each relative class, we simulated 100 replicates using the software ped-sim25, as described in Supplementary Note 8. As in a, we depict the summed length and the count of all IBD at least 12 cM long. c, Inferred IBD among four ancient English Neolithic individuals, who lived about 5,700 years ago and were entombed at Hazleton North long cairn. A full pedigree was previously reconstructed using first- and second-degree relatives inferred using pairwise SNP matching rates26. We depict all IBD at least 12 cM long. The four individuals were genotyped using 1240k aDNA capture (I12438, 3.7× average coverage on target; I12440, 2.1×; I13896, 1.1×; I12439, 6.7×).