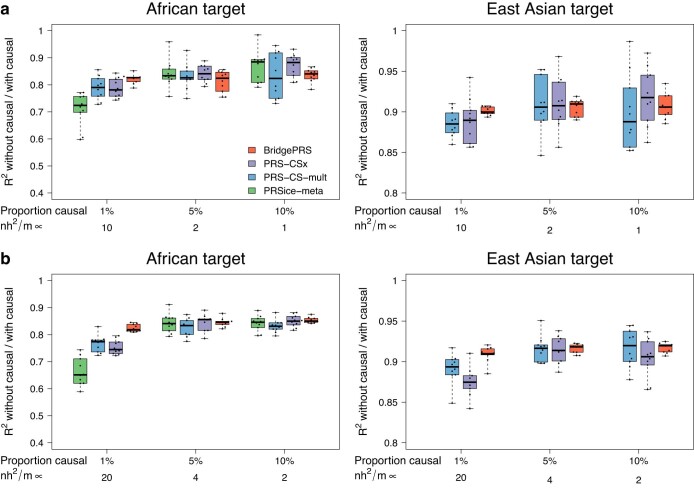
Extended Data Fig. 1. Relative loss in removing causal variants from analysis in simulated data.
Relative loss measured by ratio of models’ variance explained (R2) without and with the causal variants included. Results are shown for BridgePRS, PRS-CSx, PRS-CS-mult and PRSice-meta across six simulation scenarios for African and East Asian ancestry samples. a SNP heritability and b SNP heritability , ten simulated phenotypes per scenario. Under each set of analyses the proportion of causal variants and the relative power of the data used is shown, measured by nh2/m up to proportionality, where n is the GWAS sample size, h2 heritability and m the number of causal variants. The central rectangular boxes show the interquartile range, horizontal lines inside the boxes show the median, whiskers extend to the most extreme results and points show results for each of the 10 simulated phenotypes. PRSice-meta results for East Asian analyses were unstable and removed for clarity.