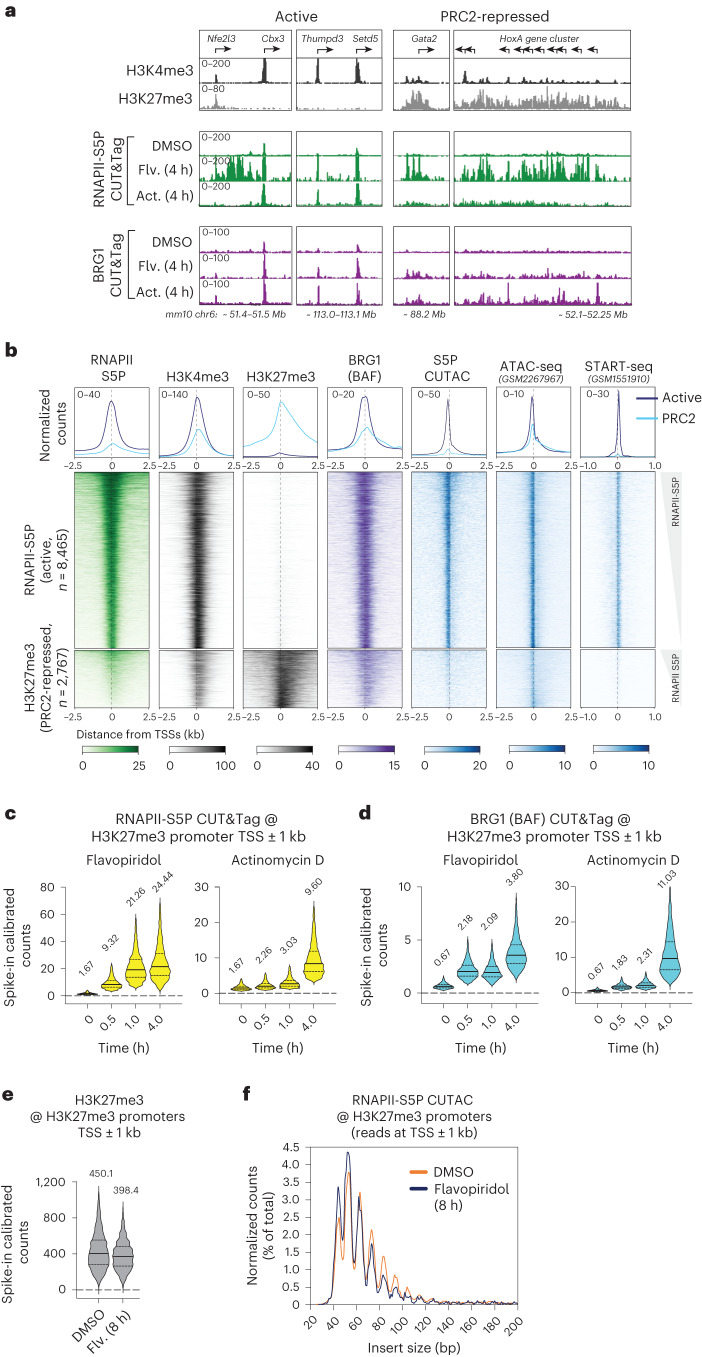
Fig. 4. Transcription inhibition shows RNAPII and BAF occupancy at Polycomb (PRC2)-repressed gene promoters.
a, Representative genomic tracks comparing enrichment of histone PTMs, RNAPII-S5P and BRG1 by CUT&Tag at transcriptionally active and PRC2-repressed genes, and changes in RNAPII-S5P and BRG1 occupancy upon Flavopiridol (Flv.) and Actinomycin D (Act.) treatment. RNAPII-S5P and BRG1 CUT&Tag read counts were spike-in calibrated. b, Heatmaps (bottom) and average plots (top) comparing histone PTMs (CUT&Tag), chromatin structure (RNAPII-S5P CUTAC and ATAC-seq) and transcriptional activity (START-seq) at RNAPII-enriched (active) and H3K27me3-enriched (PRC2-repressed) promoters. Promoters were grouped based on k-means clustering of RNAPII-S5P and H3K27me3 CUT&Tag reads mapping to a 5-kb window around the TSSs of RefSeq-annotated mESC genes (Extended Data Fig. 4a). c,d, Violin plots of spike-in calibrated CUT&Tag signal distribution comparing RNAPII-S5P (c) and BRG1 (d) occupancy over PRC2-repressed promoter TSSs ± 1 kb at time points after drug treatments. e, Violin plot comparing spike-in calibrated H3K27me3 CUT&Tag at PRC2-repressed promoter TSSs ± 1 kb in cells treated with DMSO and Flavopiridol. Median values (solid lines), upper and lower quartiles (broken lines) and outliers were calculated using the Tukey method; n = 2,767. Numbers on top of the violin plots are mean values. f, S5P CUTAC fragment size distribution to compare chromatin accessibility at PRC2-repressed promoter TSSs ± 1 kb in cells treated with DMSO and Flavopiridol. All datasets are representative of at least two biological replicates. ATAC-seq, assay for transposase-accessible chromatin using sequencing; Mb, megabase.