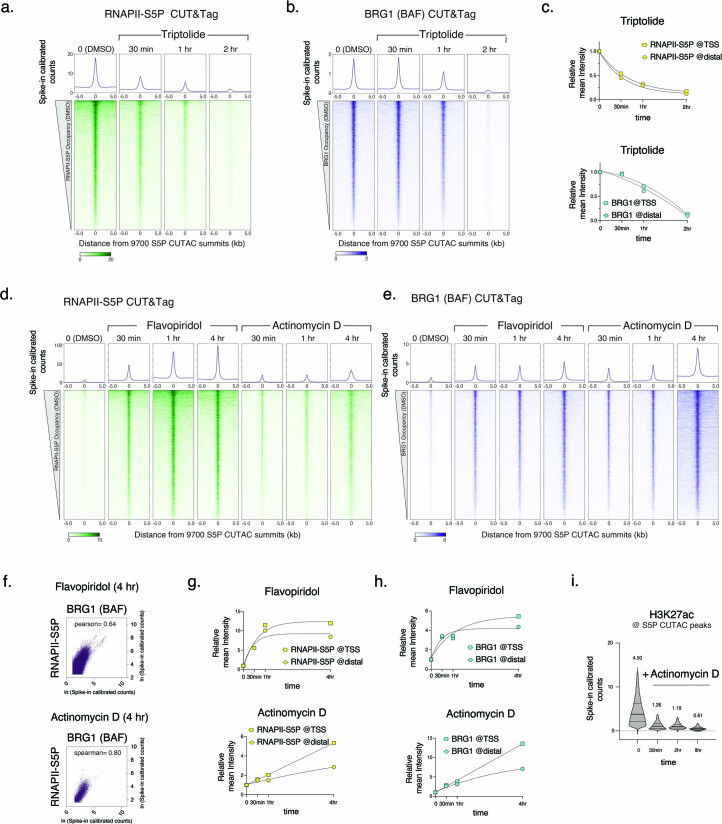
Extended Data Fig. 2. CUT&Tag of RNAPII-S5P and BRG1 after inhibitor treatment.
a, b, d, e, Heatmaps (bottom) and average plots (top) comparing RNAPII-S5P (a, d) and BRG1 (b, e) occupancy by spike-in calibrated CUT&Tag relative to the primary peaks (summits) of RNAPII-S5P CUTAC in untreated cells (DMSO) versus cells treated with Triptolide (a, b), Flavopiridol, and Actinomycin D (d, e) at indicated time points post drug treatment. c, g, h, Comparison of fold changes in mean RNAPII-S5P and BRG1 occupancy (spike-in calibrated CUT&Tag) at gene promoters (TSS, squares) and promoter-distal regulatory regions (Distal, circles) at time points after drug treatments. f, Scatterplots comparing BRG1 and RNAPII-S5P CUT&Tag reads in 1000 bp genome-wide consecutive non-overlapping bins in cells treated with Flavopiridol and Actinomycin D. i, Violin plots of CUT&Tag signal distribution comparing histone PTM H3K27ac occupancy at S5P CUTAC peaks over time points after Actinomycin D treatment. Median value (solid line), upper and lower quartiles (broken lines) and outliers were calculated using the Tukey method. Numbers on top show mean values. All datasets are representative of at least two biological replicates.