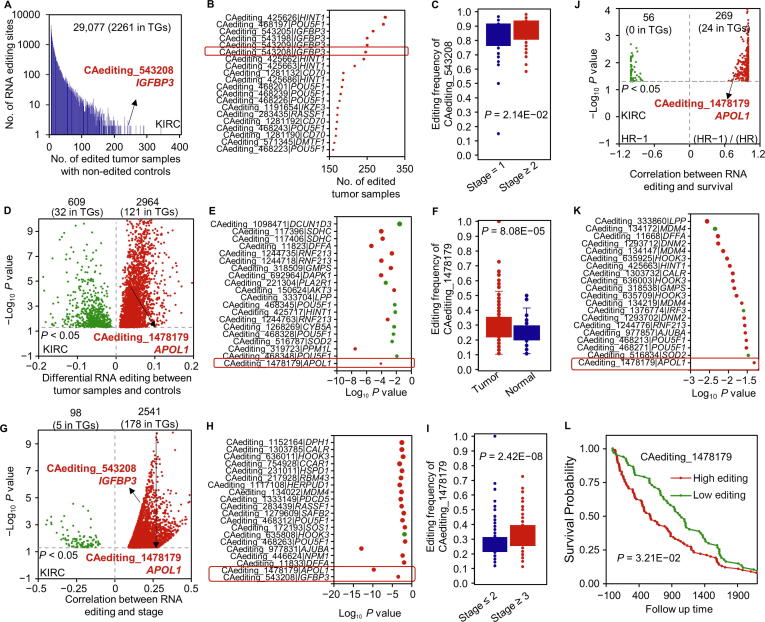
Figure 2.
RNA editing frequency analysis
A. KIRC-specific A-to-I RNA editing events with more than five edited tumor samples and non-edited normal controls. The histogram presents the distribution of this kind of RNA editing events along with the number of edited tumor samples. B. The bubble plot introduces a part of KIRC-specific RNA editing events in TGs which are not edited in normal controls. C. One significant case of IGFBP3 occurred only in 246 tumor samples for the KIRC cancer type, also showing higher editing frequencies in more severe KIRC tumors. D. KIRC-specific A-to-I RNA editing events showing differential editing frequencies in KIRC tumors compared with controls. The volcano plot presents the differences of editing frequencies between tumor samples and controls. E. The bubble plot introduces a part of KIRC-specific RNA editing events in TGs which are differentially edited in tumor samples compared with normal controls. F. One significant case in APOL1 showed higher editing frequencies in KIRC tumor samples. G. KIRC stage-associated A-to-I RNA editing events. The volcano plot presents the correlations of editing frequencies with tumor stages. H. The bubble plot introduces a part of KIRC stage-associated RNA editing events in TGs. I. One significant case in APOL1 showed higher editing frequencies in more severe KIRC tumors. J. KIRC survival-related A-to-I RNA editing events. The volcano plot presents the correlations of editing frequencies with cancer survival. K. The bubble plot introduces a part of KIRC survival-related RNA editing events in TGs. L. One significant case in APOL1 showed higher editing frequencies in the poorer survival group. The analysis results of editing frequencies for other cancer types are displayed in Figures S1–S8. KIRC, kidney renal clear cell carcinoma; TG, tumor gene; HR, hazard ratio.