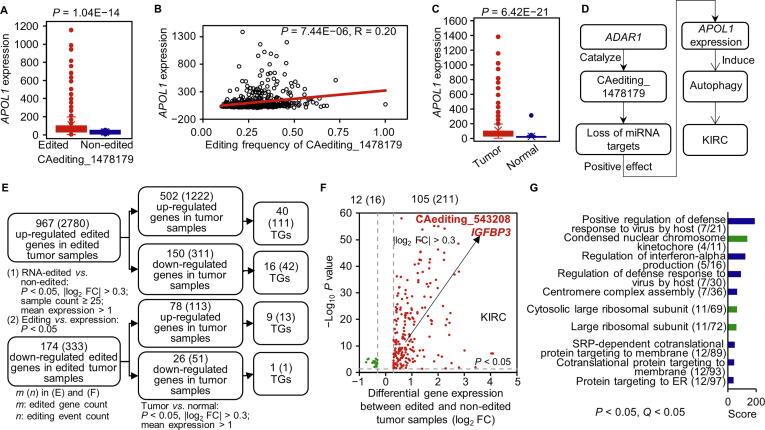
Figure 3.
The effects of A-to-I RNA editing events on gene expression
A.APOL1 was up-regulated in the RNA-edited group. B. The expression levels of APOL1 were positively associated with the frequencies of the CAediting_1478179 editing event. C.APOL1 was abnormally expressed in KIRC tumor samples compared with controls. D. The CAediting_1478179 editing event seems to be a potential biomarker for the KIRC cancer type, because it caused the loss of original miRNA binding targets to induce the up-regulated expression of APOL1, which may interfere in the autophagy function of this gene in cancer. E. The analysis procedure for the effects of A-to-I RNA editing events on gene expression. First, we performed a DEG analysis between RNA-edited and non-edited tumor samples, as well as a correlation analysis between gene expression and editing frequency to identify the DEGs whose expression levels were probably affected by A-to-I RNA editing. Then, we overlapped these genes with the DEGs identified in tumor samples compared to normal controls, to focus on RNA editing effects on the aberrantly expressed genes in cancers, especially the TGs. F. The overlapping DEGs in the KIRC cancer type. G. The overlapping DEGs were enriched in the immune- and replication-related functions and processes. The RNA editing effects on gene expression in other cancer types are presented in Figures S10–S13. FC, fold change; DEG, differentially expressed gene.