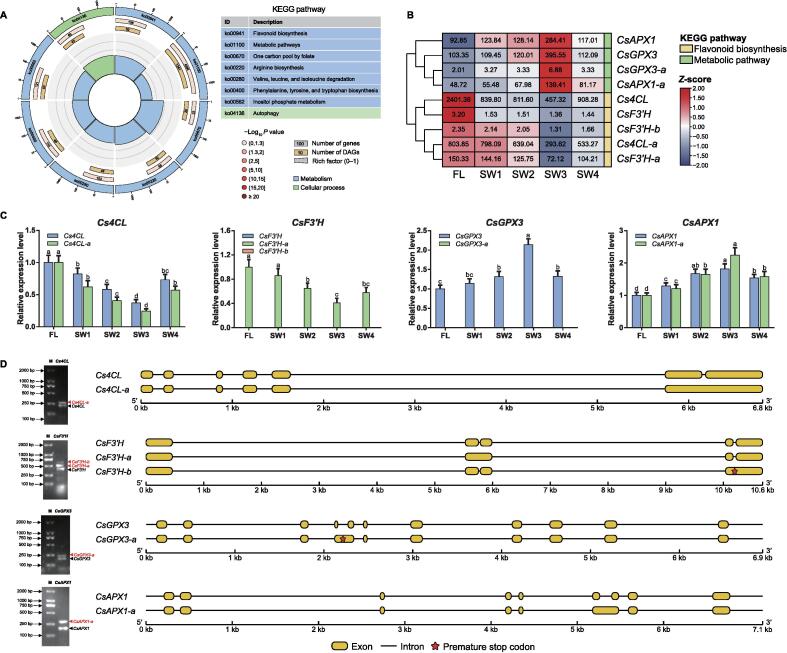
Figure 6.
AS-mediated regulatory mechanism influences the accumulation of flavor-related metabolites
A. KEGG enrichment analysis of DAGs. From the outside to the inside, the first circle indicates enriched KEGG pathways and the number of genes corresponds to the outer circle. The second circle indicates the number of genes in the genome background and the P value for the enrichment of the genes in the specified KEGG pathway. The third circle indicates the number of DAGs. The fourth circle indicates the enrichment factor of each KEGG pathway. B. The heatmap of DAGs and their AS transcripts under solar-withering with different shading rates based on the transcriptome datasets. The expression values of DAGs and their AS transcripts were normalized using the z-score formula. The number in the box indicates the original FPKM value. C. Expression patterns of Cs4CL, CsF3′H, CsGPX3, and CsAPX1 as well as their AS transcripts under solar-withering with different shading rates determined by qRT-PCR. Data are presented as mean ± SD. Different lowercase letters over the error bars indicate significantly different groups via one-way ANOVA and Tukey’s post hoc test (P < 0.05). D. RT-PCR validation and gene structure of the full-length transcripts and AS transcripts. M indicates the lane with DNA size markers. The full-length transcripts and AS transcripts on the gel images are denoted with black and red triangles, respectively. AS, alternative splicing; DAG, differentially expressed alternative splicing gene; RT-PCR, reverse transcription-polymerase chain reaction.