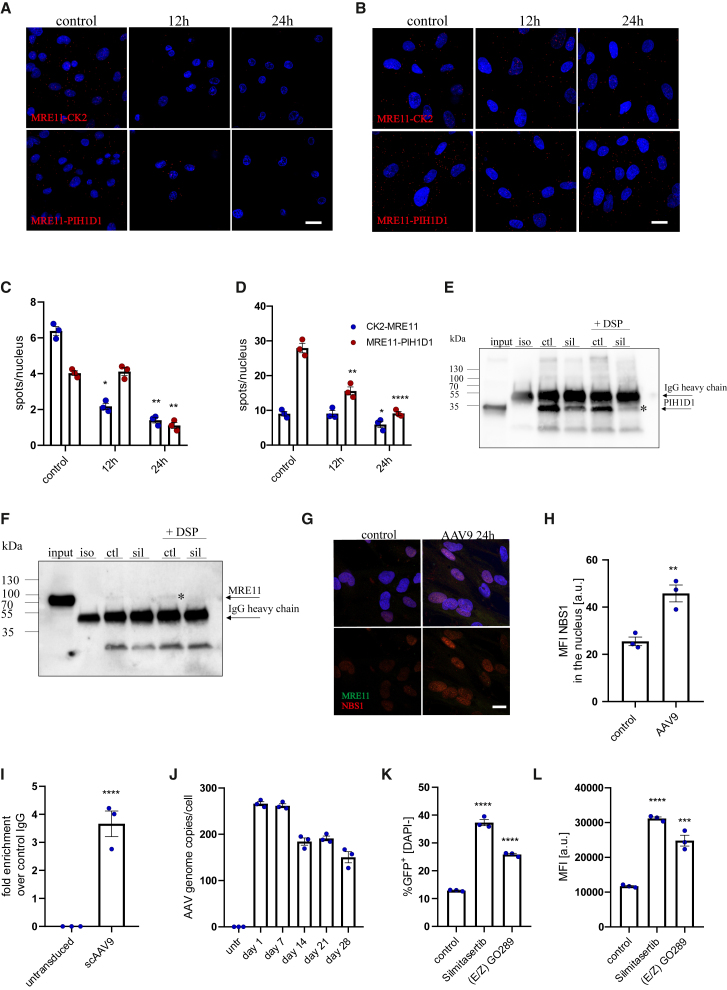
Figure 5.
Inhibition of CK2 disruptsMRE11-PIH1D1 interaction and activates transgene expression from AAV vectors
Proximity ligation assay for MRE11 and CK2 (upper panel) or MRE11 and PIH1D1 (lower panel) in iPSC-CMs (A) and iPSC-CFs (B) with quantification as number of spots per nucleus (C and D, respectively). Scale bar, 10 μm. Two-way ANOVA vs. control ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. coIP of PIH1D1 and MRE11 in iPSC-CFs with anti-PIH1D1 antibody—the membrane was probed for PIH1D1 (E) or MRE11 (F). Specific bands are marked with an asterisk. Input sample corresponds to 10% of the protein used for coIP. ctl, control; sil, cells treated for 24 h with 20 μM silmitasertib. (G) Immunofluorescent staining of MRE11 and NBS1 with MFI analysis (H) in iPSC-CFs 24 h after exposure to scAAV9-CMV-GFP vectors. Scale bar, 10 μm. (I) Quantification of genomes immunoprecipitated with anti-MRE11 antibody normalized to the signal obtained for control IgG. (J) qPCR quantification of AAV genomes in iPSC-CFs transduced with scAAV9-CMV-GFP. Analysis of transduction efficiency: percentage of GFP+ cells (K) and median fluorescence intensity (L) in iPSC-CFs treated with silmitasertib or (E/Z) GO289 assessed 5 days after scAAV9 transduction. Student’s t test vs. appropriate control ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.