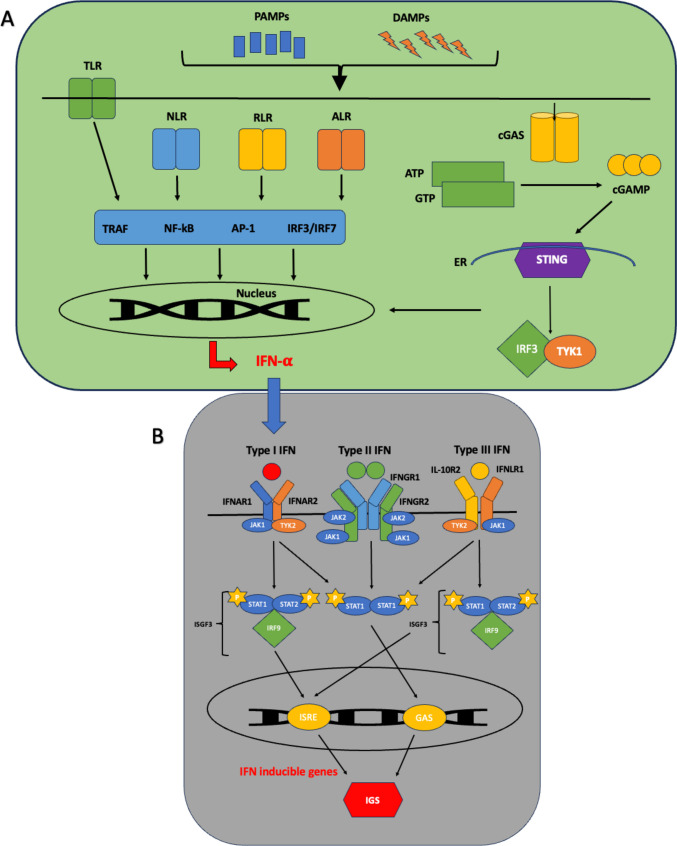
Fig. 1.
Schematic of interferon (IFN) triggers and downstream signalling pathways. A The production of IFN-I can occur following recognition of pathogen-associated molecular patterns (PAMPs), often associated with foreign bacteria or viruses, such as cytosolic DNA and double stranded RNA. These are detected by pattern recognition receptors (PRRs) which comprise of a large repertoire of germline-encoded receptors. These PRRs can be divided into subclasses including cell surface toll-like receptors (TLRs), cytosolic nod-like receptors (NLRs), retinoic acid inducible gene I receptors (RLRs), AIM2 like receptors (ALRs), and cGAS-STING pathway. Recognition of damage-associated molecular patterns (DAMPs) or PAMPS by PRRs results in transcription factor activation, such as TRAF (tumour necrosis factor receptor-associated factor), NF-kB nuclear factor kappa B, activating protein-1 (AP-1), and interferon regulatory factors (IRFs), STING (stimulator of interferon genes), and TBK1 (tank binding kinase 1), all involved in the transcription of IFN-I genes. B IFNs are categorised based on their receptor signalling, into IFN-I, IFN-II, and IFN-III. IFN-I signal via a heterodimeric receptor composed of two distinct multi-chain structures, IFN-α receptor 1 and 2 (IFNAR-1 and IFNAR-2) subunits. IFNAR associates with Janus Kinases (JAKs), with the former constitutively associated with JAK1 and the latter associated with tyrosine kinase 2 (TYK2). In response to ligand binding, these JAKs undergo activation and phosphorylate two latent transcription factors, signal transducers, and activators of transcription 1 and 2 (STAT1 and STAT2), resulting in their activation and subsequent heterodimer formation. This binds with IRF9 (IFN regulatory factor 9) or p48 to form a multi-component transcription complex called interferon-stimulated gene factor 3 (ISGF3). This complex translocates to the nucleus and binds to specific sites called IFN-stimulated response elements (ISREs), leading to the transcriptional induction of several IRGs ultimately responsible for IFN-I’s antiviral and immunomodulatory properties. The phosphorylated STAT proteins can alternatively form STAT1-STAT1 homodimers which bind gamma-activated sequences (GASs) to induce pro-inflammatory genes. As IFN-II can also signal via this alternative route (via their own heterodimeric receptor, composed of IFNGR1 and IFNGR2 subunits and associated with JAK1 and JAK 2 signalling), there can be a crossover between IFN-I and IFN-II signalling. Finally, IFN-III signals via its own heterodimeric receptor composed of IL-10R2 and IFNLR1 subunits, associated with the activation of TYK2 and JAK1, respectively. This can result in the formation and activation of STAT1-STAT2 heterodimers which associate with IRF9 to form ISGF3 complexes, with subsequent signalling as per IFN-I. AP-1, activating protein-1; DNA, deoxyribonucleic acid; ER, endoplasmic reticulum; NF-kB, nuclear factor kappa-light-chain-enhancer of activated B cells; NLR, nod-like receptor; P, phosphate; RLR, rig-I-like receptor; RNA, ribonucleic acid; TRAF, tumour necrosis factor receptor-associated factor.