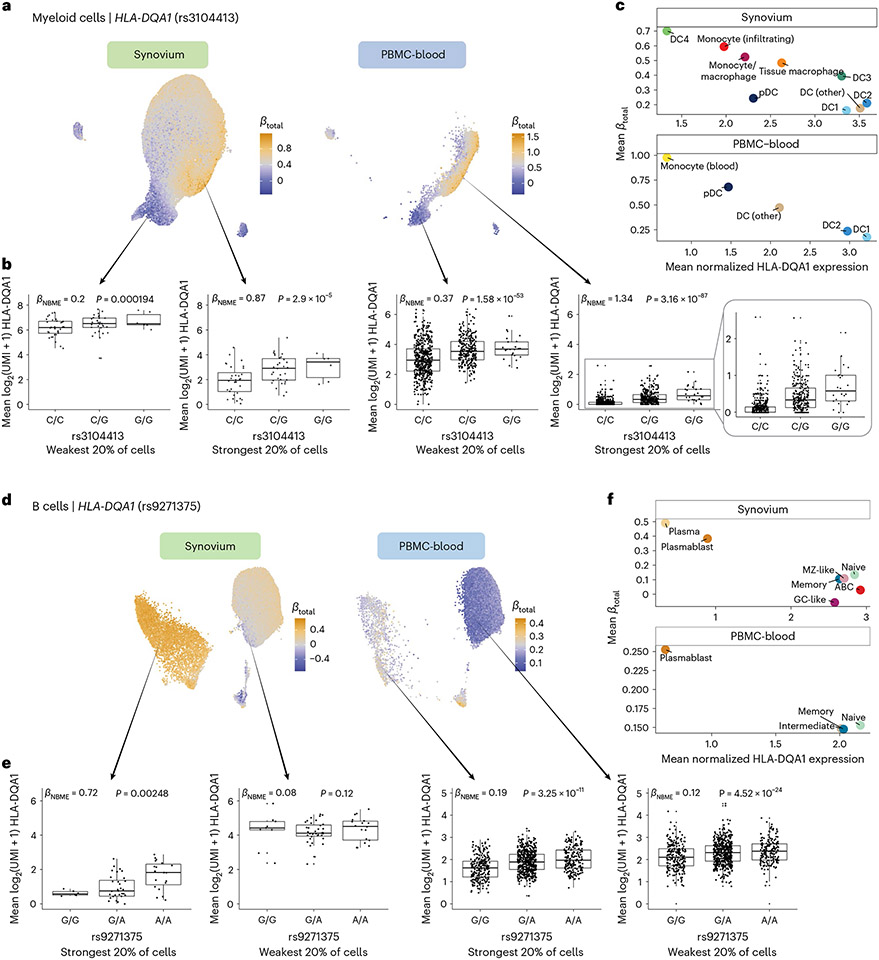
Fig. 6 ∣. Dynamic HLA-DQ eQTLs in myeloid and B cells.
a–c, Dynamic HLA-DQA1 eQTL (rs3104413) in myeloid cells (n = 69 individuals, m = 66,789 cells in synovium; n = 861, m = 40,568 in PBMC-blood). UMAP (a) of cells for tissue-defined embedding, colored by , from blue (weakest) to orange (strongest). Boxplot (b) showing the eQTL effect across individuals in the bottom and top quintiles of estimated . Labeled and P value are from fitting the NBME model without cell-state interaction terms on the cells from the discrete quintile and comparing to a null model without genotype using an LRT. Mean log2(UMI + 1) across cells per individual (y axis) by each genotype. Boxplot center line represents median, lower/upper box limits represent 25/75% quantiles, whiskers extend to box limit ± 1.5× interquartile range, and outlying points are plotted individually. Scatterplot (c) showing the mean estimated (y axis) compared to the mean log(CP10k + 1)-normalized expression of HLA-DQA1 (x axis) across annotated cell states (color). d–f, Dynamic HLA-DQA1 eQTL in B cells (n = 65 individuals, m = 25,917 cells in synovium; n = 909 individuals, m = 80,784 in PBMC-blood). d–f are analogous to a–c, respectively. LRT, likelihood ratio test (one-sided).