Figure 1.
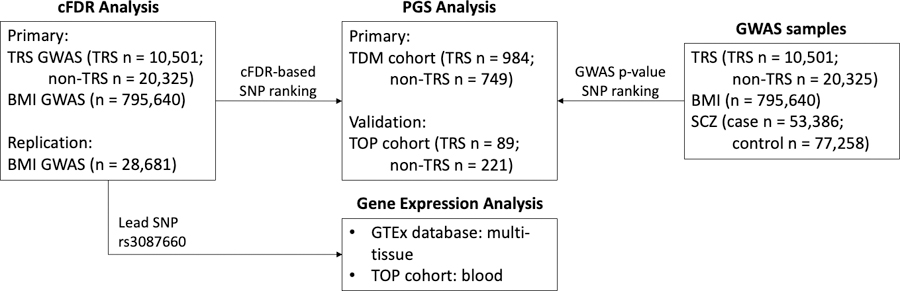
An overview of the study design and the included samples. (A) The conditional false discovery rate (cFDR) approach was used to improve discovery of genetic variants associated with treatment resistant schizophrenia (TRS) after conditioning on body-mass index (BMI). (B) Polygenic score (PGS) analyses were conducted to determine the variance in TRS explained by common genetic variants. PGSs were constructed from GWAS summary statistics of TRS, BMI and SCZ (i), as well as the re-ranked TRS data after conditioning on BMI (ii). (C) The lead SNP rs3087660, associated with TRS after conditioning on BMI, was mapped to genes using the Open Targets platform. The identified gene-variant relationships were further queried across numerous expression quantitative trait loci databases. Lasty, the identified eQTLs were investigated in the TOP TRS cohort.
