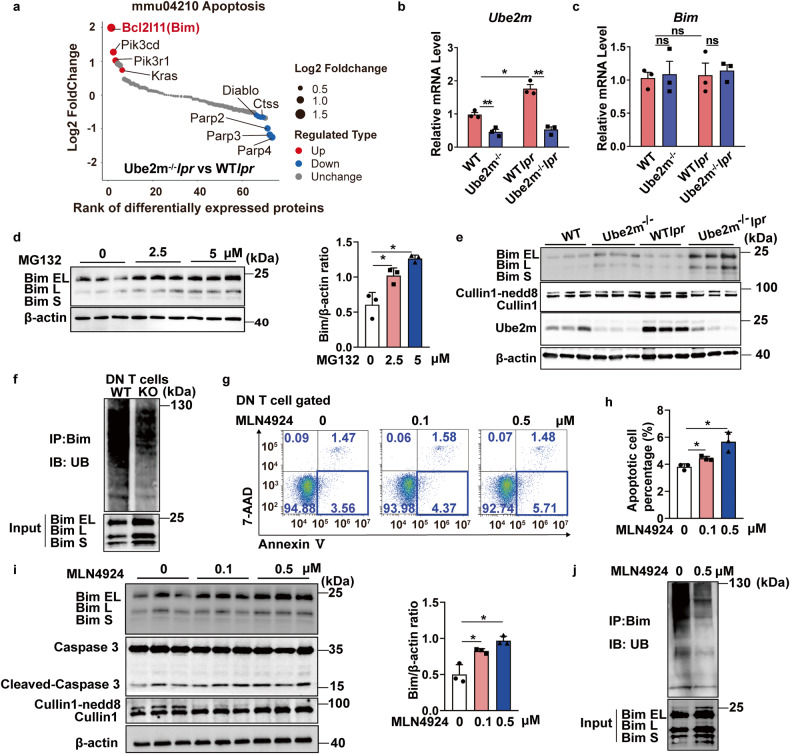
Fig. 5.
Bim level was increased with impaired ubiquitination degradation in DN T cells for neddylation inhibition. a The proteins annotated with apoptosis were selected and ranked based on the ratio value from the proteomic results of DN T cells. Red dots represented proteins significantly upregulated (FC> 1.5, P < 0.05); Blue dots represented proteins significantly downregulated (FC < 0.667, P < 0.05). n = 4/group. b, c The mRNA levels of Ube2m and Bim in DN T cells were evaluated via real-time PCR. n = 3/group. *P < 0.05, **P < 0.01. d Immunoblotting assay and densitometric analysis evaluated the protein level of Bim in DN T cells treated with MG-132. *P < 0.05. e Western blot assay was conducted to determine the level of Cullin1 neddylation, Ube2m and Bim in DN T cells. One band represented one mouse. n = 3. f Ubiquitination degradation of Bim in DN T cells was measured via Co-IP assay. Data were representative of three independent experiments. g The apoptosis of DN T cells treated with MLN4924 were detected via flow cytometry (Annexin V+/7-AAD-). h The apoptosis of DN T cells treated with MLN4924 was quantified based on the analysis of flow cytometry. n = 3/group. *P < 0.05. i Immunoblotting assay and densitometric analysis showed the level of Bim and Cleaved-caspase3 in MLN4924 treated-DN T cells. *P < 0.05. j Ubiquitination degradation of Bim in MLN4924-treated DN T cells was measured via Co-IP assay. Data were representative of three independent experiments