Table 1.
Affinities of Dihydropyridine and Pyridine Derivatives in Radioligand Binding Assays at A1, A2a, and A3 Receptorsa-f
| ||||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
|
Ki (μM) or % inhibitiond |
||||||||
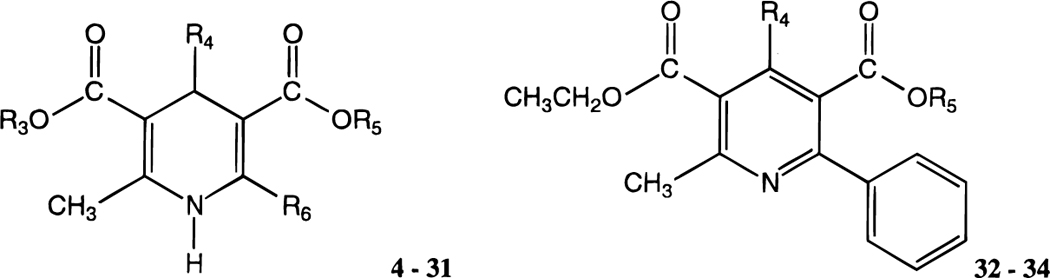
| compd | R3 | R4 | R5 | R6 | rA1a | rA2Ab | hA3c | rA1/hA3 |
|
| ||||||||
| 5 e | CH3 | CH3 | CH2CH3 | CH3 | 32.6 ± 6.3 | 46.1 ± 6.8 | 32.3 ± 5.1 | 1.0 |
| 6 e | CH3 | CH3 | CH2Ph | CH3 | 6.45 ± 1.47 | 9.72 ± 0.63 | 2.78 ± 0.89 | 2.3 |
| 7 e | CH3 | CH2CH3 | CH2CH3 | CH3 | 7.52 ± 2.79 | 7.89 ± 2.87 | 13.6 ± 2.0 | 0.53 |
| 8 e | CH3 | PhCH=CH-(trans) | CH2CH3 | CH3 | 16.1 ± 0.5 | 49.3 ± 12.5 | 0.670 ± 0.195 | 24 |
| 9 | CH2CH3 | PhCH=CH-(trans) | CH2CH3 | CH3 | 4.65 ± 1.21 | 9.23 ± 3.60 | 0.887 ± 0.138 | 5.2 |
| 10 | CH2CH3 | PhCH=CH-(trans) | CH2Ph | CH3 | 13.7 ± 2.6 | 14 ± 4% (10−4) | 3.13 ± 0.51 | 4.4 |
| 11 e | CH2CH3 | CH3 | CH2CH3 | Ph | 25.9 ± 7.3 | 35.9 ± 15.3 | 7.24 ± 2.13 | 3.6 |
| 12 | CH2CH3 | CH2CH3 | CH2CH3 | Ph | 21.5 ± 2.7 | 14.5 ± 3.5 | 8.49 ± 1.74 | 2.5 |
| 13 | CH2CH3 | CH3 | CH2Ph | Ph | 26.0 ± 8.7 | 3.15 ± 0.96 | 1.75 ± 0.47 | 15 |
| 4 e | CH2CH3 | PhCH=CH-(trans) | CH2CH3 | Ph | 5.93 ± 0.27 | 4.77 ± 0.29 | 0.108 ± 0.012 | 55 |
| 14 | CH2CH3 | PhCH=CH-(trans) | CH2CH3 | 4-CH3Ph | 14.9 ± 4.9 | 37 ± 2% (10−4) | 9.13 ± 2.43 | 1.6 |
| 15 | CH2CH3 | PhCH=CH-(trans) | CH2CH3 | 4-OCH3Ph | 9.49 ± 1.99 | d (10−4) | 1.43 ± 0.37 | 6.6 |
| 16 | CH2CH3 | PhCH=CH-(trans) | CH2CH3 | 4-ClPh | 33.0 ± 7.5 | 12 ± 7% (10−4) | 0.785 ± 0.272 | 42 |
| 17 | CH2CH3 | PhCH=CH-(trans) | CH2CH3 | 4-NO2Ph | 13.1 ± 1.6 | 40 ± 3% (10−4) | 4.14 ± 0.51 | 3.2 |
| 18 | CH2CH3 | PhCH=CH-(trans) | CH2CH3 | 3-furyl | 5.49 ± 0.25 | 35 ± 6% (10−4) | 0.907 ± 0.307 | 6.1 |
| 19 | CH2CH3 | PhCH=CH-(trans) | CH2CH3 | 3-thienyl | 7.52 ± 1.38 | d (10−4) | 0.407 ± 0.066 | 18 |
| 20 | CH2CH3 | 2-OCH3PhCH=CH-(trans) | CH2CH3 | Ph | 25.3 ± 2.4 | 17 ± 12% (10−4) | 0.334 ± 0.059 | 76 |
| 21 | CH2CH3 | 2-NO2PhCH=CH-(trans) | CH2CH3 | Ph | 6.03 ± 1.39 | d (10−4) | 0.109±0.017 | 55 |
| 22 | CH2CH3 | 4-NO2PhCH=CH-(trans) | CH2CH3 | Ph | 23 ± 9% (10−4) | 33% (10−4) | 0.0585 ± 0.0164 | >1700 |
| 23 | CH2CH3 | 4-NH2PhCH=CH-(trans) | CH2CH3 | Ph | 31 ± 3% (10−4) | 26 ± 6% (10−4) | 0.198 ± 0.047 | >500 |
| 24 | CH2CH3 | (Ph)2C=CH- | CH2CH3 | Ph | 1.20 ± 0.14 | 12 ± 6% (10−4) | 1.42 ± 0.23 | 0.84 |
| 25 | CH2CH3 | PhC≡C- | CH2CH3 | Ph | 11.0 ± 0.1 | 26 ± 12% (10−4) | 0.0766 ± 0.0151 | 140 |
| 26 | CH2CH3 | PhCH=CH-(trans) | CH2Ph | Ph | 35 ± 3% (10−4) | 15 ± 3% (10−4) | 0.0583 ± 0.0124 | >1700 |
| 27 | CH2CH3 | 4-NO2PhCH=CH-(trans) | CH2Ph | Ph | 33 ± 1% (10−4) | d (10−4) | 0.0724 ± 0.0377 | >1300 |
| 28 | CH2CH3 | PhC≡C- | CH2Ph | Ph | 40.1 ± 7.5 | d (10−4) | 0.0314 ± 0.0028f | 1300 |
| 29 | CH2Ph | PhCH=CH-(trans) | CH2CH3 | Ph | d (10−4) | 16 ± 11% (10−4) | 0.142 ± 0.047 | >700 |
| 30 | CH2Ph | 4-NO2PhCH=CH-(trans) | CH2CH3 | Ph | d (10−4) | d (10−4) | 0.286 ± 0.038 | >400 |
| 31 | CH2Ph | PhC≡C- | CH2CH3 | Ph | 24 ± 4% (10−4) | d (10−4) | 0.169 ± 0.026 | >600 |
| 32 e | CH3 | CH2CH3 | 7.41 ± 1.29 | 28.4 ± 9.1 | 4.47 ± 0.46 | 1.7 | ||
| 33 | PhCH=CH-(trans) | CH2CH3 | 2.49 ± 0.47 | 2.40 ± 0.22 | 2.80 ± 1.78 | 0.85 | ||
| 34 | PhC≡C- | CH2Ph | 11.6 ± 4.8 | 43 ± 2% (10−4) | 2.75 ± 0.78 | 4.2 | ||
Displacement of specific [3H]-(R)-PIA binding in rat brain membranes, expressed as Ki ± SEM in μM (n = 3–5), or as a percentage of specific binding displaced at the indicated concentration (M).
Displacement of specific [3H]CGS 21680 binding in rat striatal membranes, expressed as Ki ± SEM in μM (n = 3–6), or as a percentage of specific binding displaced at the indicated concentration (M).
Displacement of specific [125I]AB-MECA binding at human A3 receptors expressed in HEK cells, in membranes, expressed as Ki ± SEM in μM (n = 3–4).
Displacement of ≤ 10% of specific binding at the indicated concentration (M).
Values taken from van Rhee et al.16
Ki value at rat A3 receptors stably expressed in CHO cells1 found to be 3.53 ± 0.61 μM.
