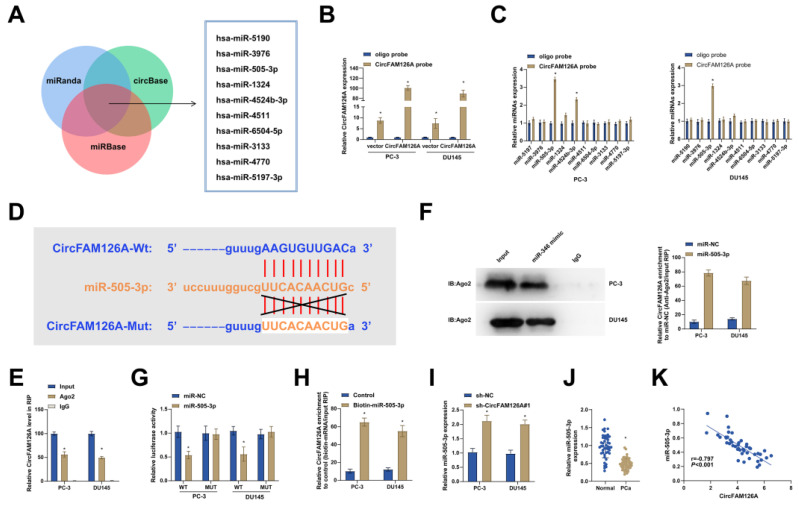
Figure 5.
circFAM126A as a sponge for miR-505-3p. A: The potential miRNAs of CircFAM126A were predicted using bioinformatics websites such as miRanda, miRBase, and circBase. B: Lysates of PCa cells overexpressing CircFAM126A were subjected to a biotinylated pull-down assay, and the expression levels of CircFAM126A were detected by RT-qPCR. C: Candidate miRNAs of circFAM126A were analyzed using the biotinylated pull-down assay. D: The bioinformatics website Starbase predicted potential binding sites between CircFAM126A and miR-505-3p. E and F: Ago2-RIP assays confirmed the targeting relationship between miR-505-3p and circFAM126A. G: Dual-luciferase reporter assays were performed to investigate the targeting relationship between miR-505-3p and circFAM126A. H: The targeting relationship between miR-505-3p and circFAM126A was analyzed through a biotinylated pull-down assay. I: The impact of CircFAM126A knockdown on the expression of miR-505-3p was determined by RT-qPCR. J: The expression of miR-505-3p in cancer tissues and adjacent normal tissues was measured by RT-qPCR. K: Pearson correlation analysis indicated a negative correlation between CircFAM126A and miR-505-3p levels in the tumor tissues of 46 PCa patients. Data are represented as mean ± SD (n = 3), with * P < 0.05 denoting statistical significance.