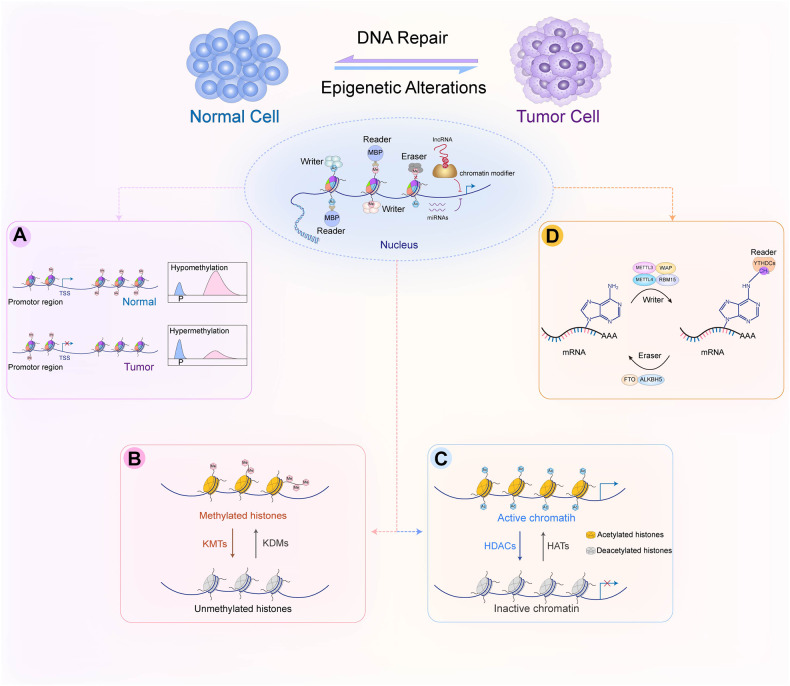
Fig. 1. Epigenetic alterations associated with carcinogenesis.
Epigenetic alterations involve DNA methylation, histone acetylation, and miRNA regulation that have reversible effects on gene silencing and activation through epigenetic enzymes and related proteins. Writers (DNMT, HAT, and KMT) are enzymes that add acetyl (Ac) and methyl (Me) tags to histones. MBDs are readers that recognize methyl-CpG and modify histones. Erasers (DNA demethylase, HDAC, and KDM) are responsible for removing chemical groups from DNA or histones. Noncoding RNAs (miRNAs and lncRNAs) are also involved in epigenetic regulation. A DNA methylation in normal and cancer cells. The overall hypomethylation and local hypermethylation of promoter regions are characteristics of cancer cells. P: promoter region. B Methylation and demethylation of lysine or arginine in histones. Lysine can be methylated once (me1), twice (me2) or three times (me3) catalyzed by KMT. Arginine is methylated once (me1) or twice (me2) catalyzed by KMT. These processes can be reversed by KDM. C HDAC removes acetyl groups from histone lysine residues. Acetylated histones are considered “active chromatin” allowing gene transcription, whereas deacetylated histones are “non-active chromatin” associated with gene silencing. D The methylation of m6A is installed by the RNA methyltransferase complex with the catalytic subunit METTL3/METTL4 (writer) and removed by demethylases, such as FTO and ALKBH5 (eraser). m6A reader proteins (YTHDCs) can specifically bind m6A transcripts. DNMT, DNA methyltransferase; HAT, histone acetyltransferase; HDAC, histone deacetylase; KDM, lysine demethylase; KMT, lysine methyltransferase; m6A, N6-Methyladenosine. MBP, methyl-CpG-binding domain protein.