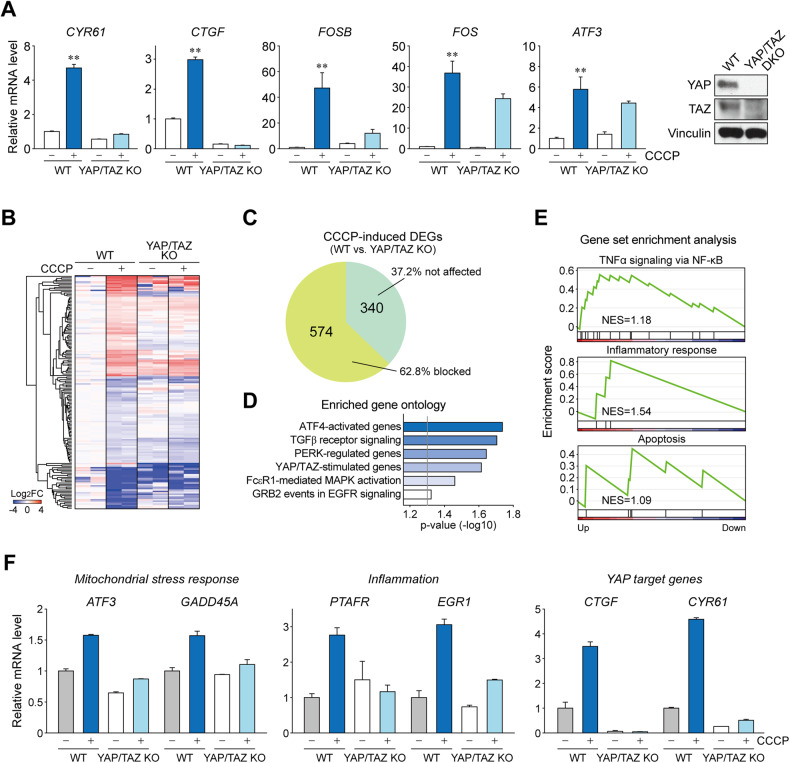
Fig. 6. YAP/TAZ induce the mitochondrial stress transcriptome.
A qRT-PCR analysis for YAP/TAZ target genes were performed using YAP/TAZ KO and wild-type (WT) after CCCP treatment (n = 3 per group). B RNA-seq analysis of YAP/TAZ KO and wild-type cells was performed and heatmap of CCCP-induced DEGs which are affected by YAP/TAZ KO is shown (n = 2 per group). Cells were harvested 3 h after CCCP (20 μM) treatment. C Pie chart showing the portion of CCCP-induced DEGs blocked by YAP/TAZ KO. D Gene ontology analysis for YAP/TAZ-dependent DEGs. The genes were categorized according to the KEGG pathway. E Gene set enrichment analysis of the DEGs shows enriched TNFα, inflammation, and apoptosis signaling pathways, respectively. DEGs were grouped according to the Hallmark gene sets in molecular signature database (MSigDB). F Expression of genes related to mitochondrial stress response and inflammation and YAP/TAZ transcriptional targets from the RNA-seq experiment.