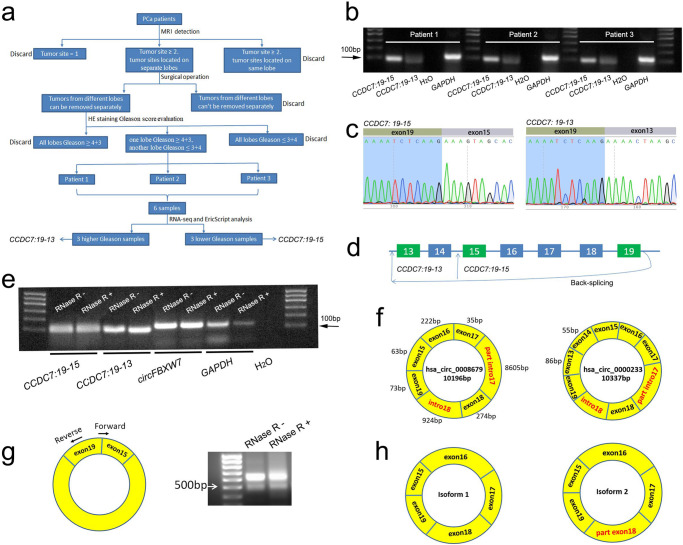
Fig. 1. The discovery of circCCDC7(15,16,17,18,19) in PCa.
a The pipeline for discovering prostate cancer chimeric RNAs: 1, based on MR reports, we focused on patients having tumor sites on two separate lobes; 2, during the surgery, the target tumors are dissected according to MR images; 3, half of the tumor was submitted to pathologist to receive a Gleason score evaluation for each tumor site; 4, selected tumors from the same patients with one lobe’s Gleason score higher than 4 + 3, and another lower than 3 + 4; 5, six tumor tissues from three patients were obtained and submitted for RNA-Seq. Finally, EricScript software was performed to predict chimeric RNAs. b Gel image of RT-qPCR product of CCDC719-15 and CCDC719-13 in three clinical patients. Mix cDNA from tumor and adjacent normal margin was used. c Sanger sequencing results of the validated CCDC719-15 and CCDC719-13. d The schematic diagram of CCDC719-15 and CCDC719-13. e Confirmation for CCDC719-15 and CCDC719-13 both being circular RNAs which circFBXW7 was used as a control. Total mix RNA from 23 pairs PCa patients was treated with or without RNase R before performing RT-qPCR. f The structures of two reported circular RNAs which share the same junction sequences with CCDC719-15 and CCDC719-13. g Gel image of Touch-down PCR products of CCDC719-15 using divergent primers. The complementary DNA was synthesized with random hexamer primer using total mix RNA that from 23 pairs PCa patients treated with or without RNase R. h The schematic diagram of different isoforms of circCCDC7.