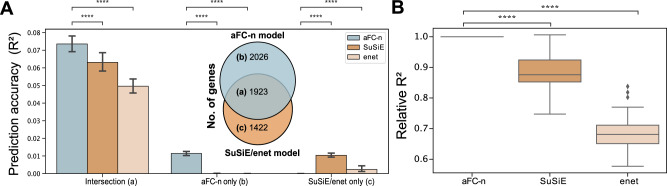
Fig. 3. The aFC-n improves the predictive accuracy of genetically regulated gene expression.
Comparing predicted gene expression from the aFC-n, elastic net (enet)27 and SuSiE28 models, using out of sample data. For SuSiE and enet we considered genes with significant expression cis-heritability. For aFC-n we considered the model for genes with eQTLs. A The aFC-n model outperforms SuSiE and elastic net for 1923 genes shared by all models (subpanel a) in adipose subcutaneous tissue (median R2 is 0.073 for the aFC-n model and 0.063 for the SuSiE predictive model; The two-sided Wilcoxon signed-rank test p value: 1.8 × 10−47). For the genes not shared between models, the performance is much lower for all models (subpanels b, c). Error bars represent 95% bootstrap confidence intervals of the median. B The distribution of the median R2 relative to the median of the R2 for aFC-n model for 47 tissues for shared genes (two-sided wilcoxon signed rank test p value = 2.6 × 10−9 and 2.4 × 10−9,comparing aFC-n with SuSie and enet, respectively). Comparing aFC-n with SuSiE and elastic net prediction models, Wilcoxon signed-rank tests are significant (FDR < 0.05) for 46 and 47 tissues, respectively. The p value annotation: ****p ≤ 10–4. Boxplots represent first quartile, median, and third quartiles. Whiskers represent Q1–1.5* interquartile range (IQR) and, Q3 + 1.5*IQR.