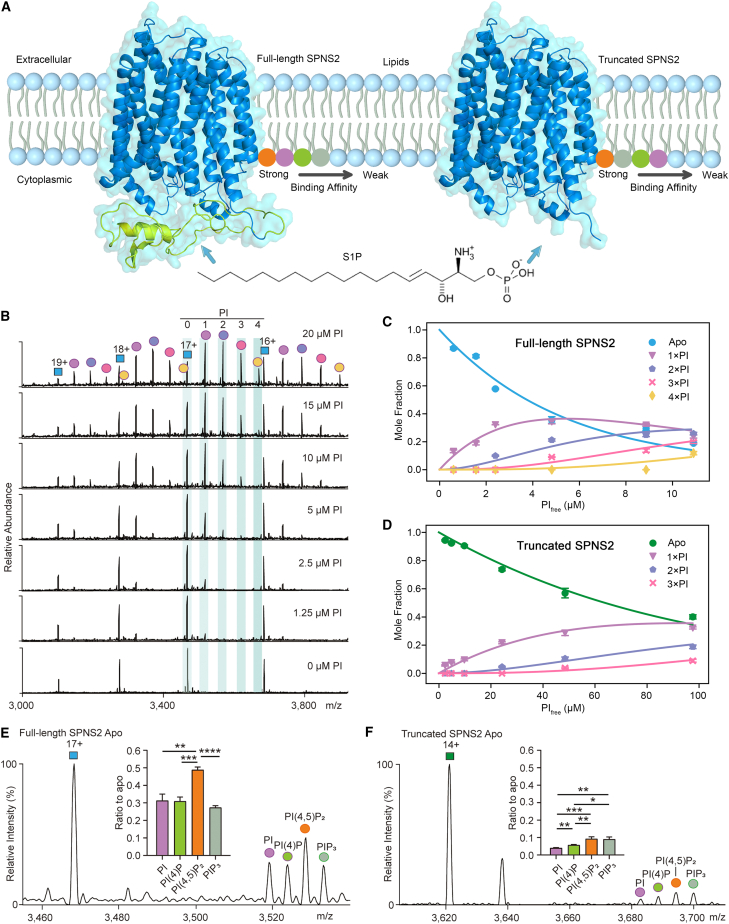
Figure 1.
A predicted model of SPNS2 and the binding preference of PI derivatives to full-length and truncated SPNS2
(A) The AlphaFold2 predicted model of SPNS2 (blue) with the N terminus of the full-length protein (green) shown schematically, embedded within a phospholipid bilayer. PI derivatives: PI, PI(4)P, PI(4,5)P2, and PIP3 are shown as purple, green, orange, and gray spheres, respectively, with binding preference to full-length and truncated SPNS2 arranged via proximity to the transporter (according to the KD values in Table S1). The structure of S1P is also shown.
(B) Mass spectra recorded for delipidated full-length SPNS2 (5 μM) with increasing concentrations from 0 to 20 μM of PI (18:1/18:1).
(C) Plot of the mole fraction of full-length SPNS2 and PI binding calculated from titration of PI (18:1/18:1) with a resulting fit (R2 = 0.97) from a sequential ligand-binding model (solid lines). Data are plotted as mean ± standard deviation (SD) (n = 3).
(D) Plot of mole fraction of truncated SPNS2 with PI-bound states calculated from titration of PI(18:1/18:1) with a resulting fit (R2 = 0.99) from a sequential ligand-binding model (solid lines). Data are plotted as mean ± SD (n = 3).
(E) A single charge state (17+) of full-length SPNS2 following incubation with an equimolar solution containing PI, PI(4)P, PI(4,5)P2, and PIP3 confirms preferred binding of PI(4,5)P2. Data are plotted as mean ± SD (n = 3). ∗∗p < 0.01. ∗∗∗p < 0.001. ∗∗∗∗p < 0.0001.
(F) A single charge state (14+) of truncated SPNS2 following incubation with an equimolar solution containing PI, PI(4)P, PI(4,5)P2, and PIP3. Data are plotted as mean ± SD (n = 3). ∗p < 0.05. ∗∗p < 0.01. ∗∗∗p < 0.001.