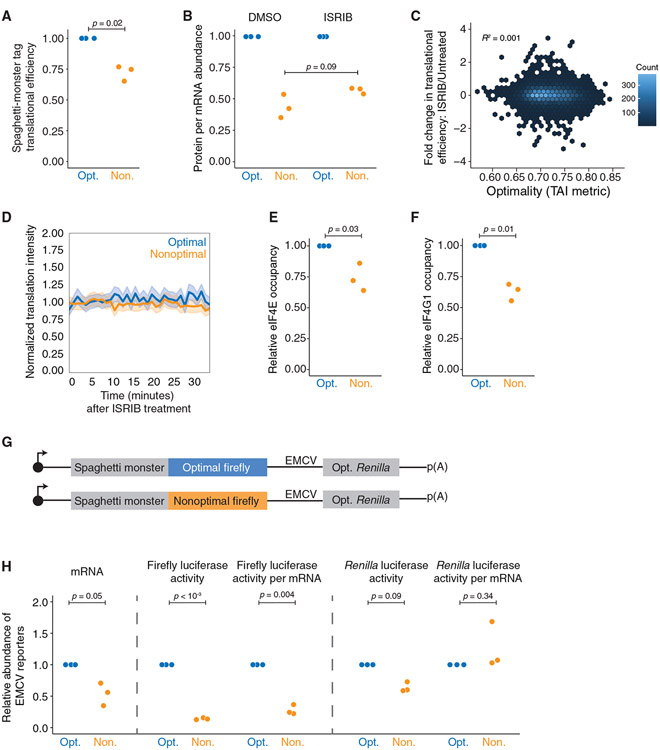
Figure 5. Nonoptimal codons inhibit translation initiation through reduced eIF4E and eIF4G1 binding.
(A) Nonoptimal codons reduce ribosome association upstream. Shown is the normalized translational efficiency for the N-terminal SM tag, which is upstream of the optimal or nonoptimal firefly luciferase ORF, in the dual-tagged reporters (n = 3).
(B) ISRIB does not rescue the translational efficiency of the nonoptimal reporter. HEK293T cells were transfected with plasmids expressing each reporter alongside optimized Renilla luciferase. Shown are normalized protein-per-mRNA abundance, mRNA abundance (RT-qPCR), and protein activity (dual-luciferase assay); n = 3.
(C) ISRIB does not rescue the translational efficiency throughout the human transcriptome in an optimality-dependent manner. Meta-analysis was performed from published Ribo-seq data. Shown is the change in translational efficiency of the HEK293T transcriptome when unstressed cells were treated with ISRIB, plotted against their tAI optimality score (Rep “A”).
(D) ISRIB does not rescue the number of ribosomes translating nonoptimal SM-tagged luciferase reporter transcripts. Plotted is the average change in translational intensity on individual reporter transcripts, tracked over time in U2OS cells, following DMSO or ISRIB treatment. For the nonoptimal reporter, 17 cells were imaged (338 total translation spots). For the optimal reporter, 13 cells were imaged (242 total translation spots). Mean values ± SD are shown.
(E) Nonoptimal codons reduce eIF4E binding. HEK293T cells were transfected with either optimal or nonoptimal SM-tagged firefly luciferase reporters, along with Renilla luciferase, and then eIF4E RNA immunoprecipitations (RIPs) were performed (n = 3).
(F) Nonoptimal codons reduce eIF4G1 binding. As in (E) except for eIF4G1 RIPs (n = 3).
(G) Schematic of reporter constructs with Renilla luciferase being expressed by an EMCV IRES.
(H) eIF4E is required to reduce translation initiation on nonoptimal transcripts. HEK293T cells were transfected with plasmids expressing each reporter, along with γHA-GFP as a transfection control. mRNA abundance was quantified by RT-qPCR, with values normalized to GFP. Protein activity was quantified by dual-luciferase activity. Shown are the fold changes in the nonoptimal reporter relative to the optimal: (1) (left) mRNA abundance, (2) (middle) firefly luciferase activity, and firefly luciferase activity relative to mRNA abundance, and (3) (right) Renilla luciferase activity, and Renilla luciferase activity relative to mRNA abundance; n = 3.