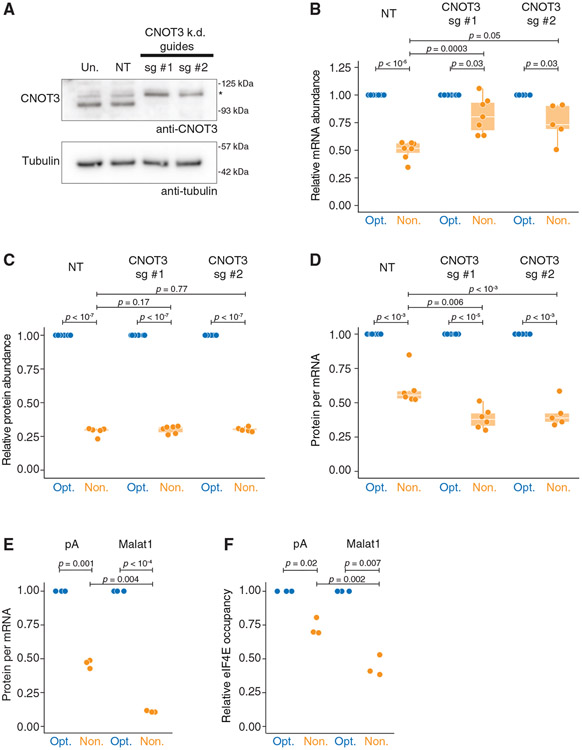
Figure 6. Translation repression does not require mRNA decay or deadenylation.
(A) CRISPR guides can effectively knock down CNOT3. HEK293T lysates were collected 4 days following blasticidin selection and probed for CNOT3 (105 kDa expected; *, nonspecific band) and tubulin.
(B) Upon CNOT3 knockdown, nonoptimal mRNA levels are rescued relative to optimal. Following 4 days of blasticidin selection, cells were transfected with SM-firefly luciferase reporters and optimized Renilla luciferase. RT-qPCR quantified mRNA abundance. Box-and-whisker plots show relative fold change in transcript (guide #1: n = 7; guide #2: n = 5).
(C) Upon CNOT3 knockdown, nonoptimal protein activity is unchanged. Like in (B) but measured relative protein levels by dual-luciferase assay (guide #1: n = 7; guide #2: n = 5).
(D) Upon CNOT3 knockdown, there is more translational repression of the nonoptimal reporter. Values plotted are extrapolated from (B) and (C), calculating protein activity relative to transcript abundance.
(E) Neither deadenylation nor a poly(A) tail is required for translational repression. HEK293T cells were transfected with plasmids expressing optimality reporters, which, after processing, ended in a poly(A) tail or Malat1 triple helix. Shown is a dot plot depicting normalized protein-per-mRNA abundance (dual-luciferase assay and RT-qPCR; n = 3).
(F) Without a poly(A) tail, nonoptimal codons further reduce eIF4E binding. HEK293T cells were transfected with plasmids as in (E), and eIF4E RIPs were performed; n = 3.