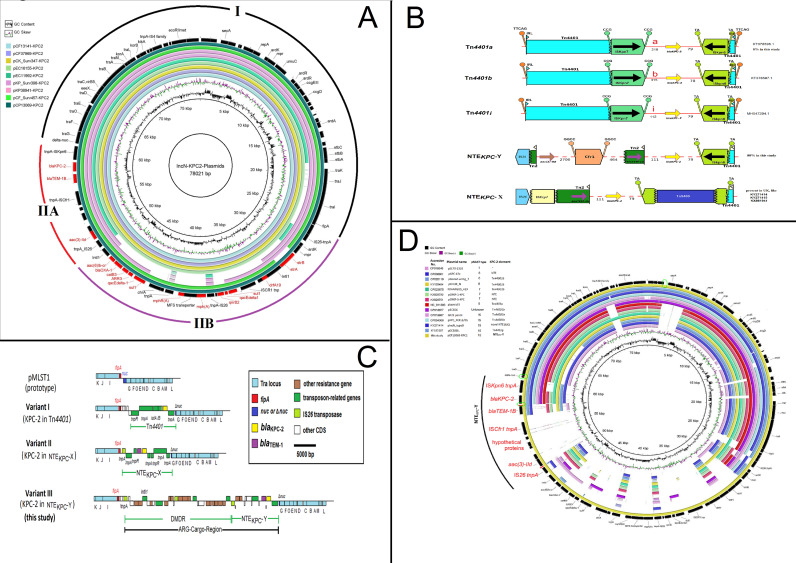
Fig 2.
(A) Genetic map of the representative complete IncN[pMLST15] plasmids with the common length of 78,021 bp depicting the backbone (region I), the unique genetic environment NTEKPC-Y (region IIA), and the dynamic multi-drug resistance region (DMDR; Region IIB). Antimicrobial resistance genes (ARGs) are marked in red. The genetic environment of the NTEKPC-Y (region IIA), encoding bla KPC-2–bla TEM-1B and acc (3)IId, was present in all plasmids; the DMDR (region IIB) differed in some plasmids, representing different antimicrobial resistance profiles. (B) Comparison of the non-Tn4401-KPC-element NTEKPC-Y (this study), NTEKPC-X (from United Kingdom), and Tn4401 variants from the KPC-2-encoding IncN[pMLST15] plasmids. (C) Insertion sites of the ARG-bearing cargo-region of different IncN[pMLST15] variants. Variant I, bla KPC-2 located in Tn4401 (CP004366, CP004367, CP018963, KX154765, KX276209, KX062091, KX397572, KF182187, and KF181264); variant II, bla KPC-2 located in and NTEKPC-X (KX88194, KY27414, and KY27415); and variant III, the bla KPC-2, was located in NTEKPC-Y (this study). (D) Comparison of different KPC-2 IncN pMLST types. The pCP13069-KPC2 contained a unique region, assigned as NTEKPC-Y. The green circles on the ring maps symbolize the areas for the specific PCR primers.