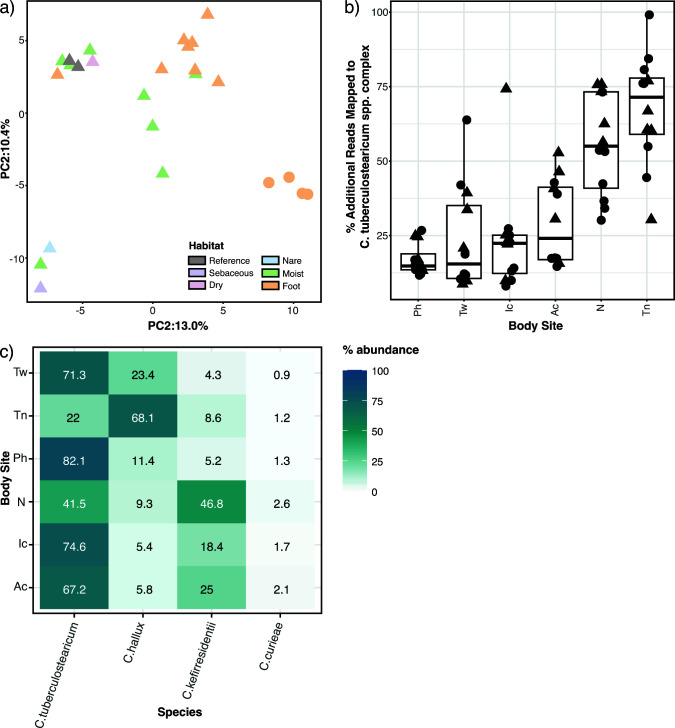
Fig 4.
C. tuberculostearicum complex pangenome clustering and improved metagenomic read mapping. (a) PCA of orthologous gene clustering. The gene presence/absence data for 25 genomes (including two NCBI references, shown in gray) was analyzed using PCA. Ribotype B genomes are shown as circles; other genomes are triangles. (b) Improvement in shotgun metagenomic read mapping with a 28 member C. tuberculostearicum database as compared to the five member NCBI database. Percent increase in mapped C. tuberculostearicum reads by body site. Each point is an HV. Triangles mark HVs that contributed one or more isolates to the expanded mapping database. (c) Relative abundance of C. tuberculostericum species complex members, including the newly proposed species C. hallux, based on bracken-corrected kraken2 analysis.