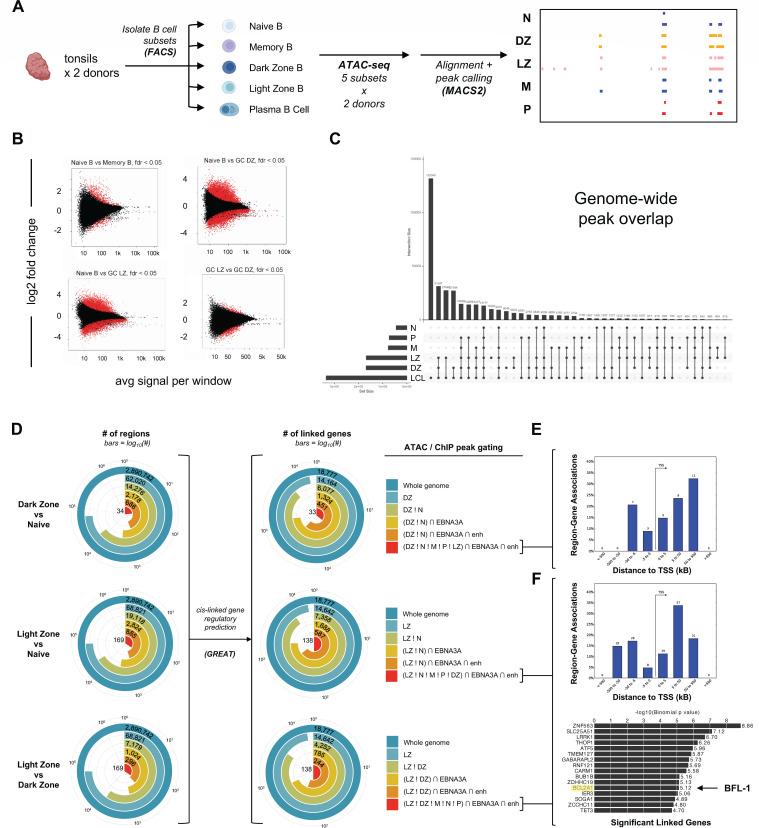
Fig 3.
Identification of open chromatin regions in tonsillar B cells and prediction of genes linked to differentially accessible EBNA3A enhancer loci. (A) Schematic of ATAC-seq experiment indicating that data were generated from donor-matched CD19+ B cells sorted for naïve, memory, GC DZ, GC LZ, and plasma cells (PC). (B) MA plots (log2 fold change vs. mean average) showing differentially accessible regions between fractions of interest from tonsils. Red points indicate regions with a false discovery rate (FDR) <0.05. (C) Upset plot of genome-wide overlapping accessible chromatin regions among tonsillar B-cell subsets and LCLs. (D) Bullseye plots depicting the fraction of intervals with differential accessibility among tonsillar subsets, their co-incidence with EBNA3A and enhancer ChIP-seq signatures in LCLs, and cis-linked gene predictions. Logical gates were applied to filter ATAC peaks present in one subset but not in others (e.g., LZ ! DZ) and to intersect these differentially accessible loci against ChIP-seq peaks for EBNA3A and histone enhancer patterns (e.g., ∩ EBNA3A and/or ∩ enh). Regions matching given logical gating criteria were filtered to exclude any intervals shorter than 100 bases. Genes with predicted cis-regulatory linkages to intervals matching each set of gates were identified using GREAT (44). (E) Distribution of DZ-unique EBNA3A- and enhancer-associated gene-linked intervals relative to gene TSSs. This gate recipe [(DZ ! LZ ! N ! M ! P) ∩ EBNA3A ∩ enh] yielded no significant linked genes. (F) Distribution of LZ-unique EBNA3A- and enhancer-associated gene-linked intervals relative to gene TSSs. This gate recipe [(LZ ! LZ ! N ! M ! P) ∩ EBNA3A ∩ enh] yielded multiple statistically significant linked genes (binomial gene test FDR Q <0.05), including BCL2A1 (BFL-1).