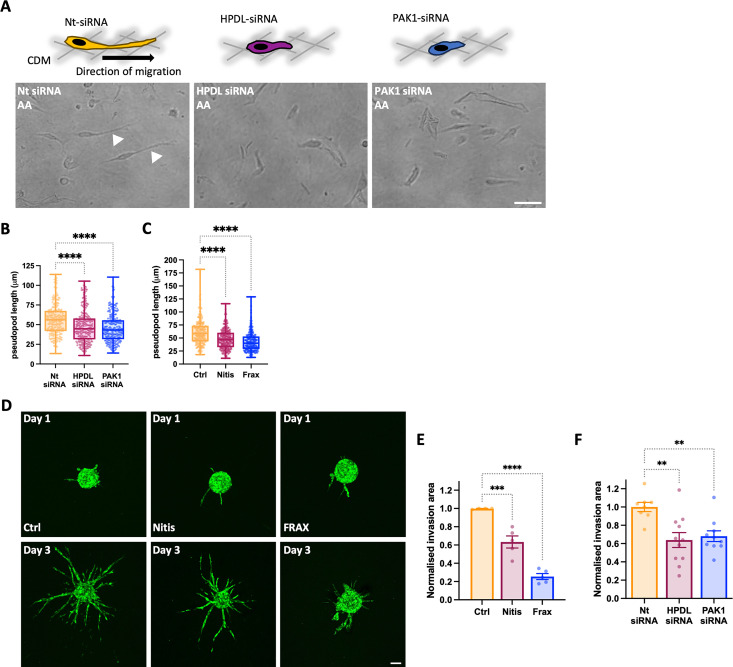
Fig 8. Tyrosine catabolism supported cell migration and invasion in 3D systems.
(A–C) MDA-MB-231 cells, transfected with siRNA targeting PAK1 (PAK1 siRNA), HPDL (HPDL siRNA), or a non-targeting siRNA control (Nt siRNA) when indicated, where plated on CAF-CDM for at least 4 h, treated with 3 μm FRAX597 (FRAX) or 40 μm Nitisinone (Nitis) and imaged for 16 h by time lapse microscopy. Arrowheads indicate elongated pseudopods. Bar, 30 μm. Pseudopod elongation was measured with Image J. Values are from 3 independent experiments, ****p < 0.0001 Kruskal–Wallis, Dunn’s multiple comparisons test. (D–F) GFP-MDA-MB-231 cell spheroids were generated by the hanging drop method, embedded in 3 mg/ml collagen I and geltrex (50:50) mixture. Cells were transfected or spheroids were treated as in A–C, starved of amino acid and imaged live with a Nikon A1 confocal microscope. Invasion area was calculated with Image J, by subtracting the core area from the total spheroid area. Values are mean ± SEM from 3 independent experiments, **p < 0.01; ***p < 0.001; ****p < 0.0001 Kruskal–Wallis, Dunn’s multiple comparisons test. All the raw data associated with this figure are available in S9 Data. CAF, cancer-associated fibroblast; CDM, cell-derived matrix; HPDL, p-hydroxyphenylpyruvate hydroxylase-like protein.