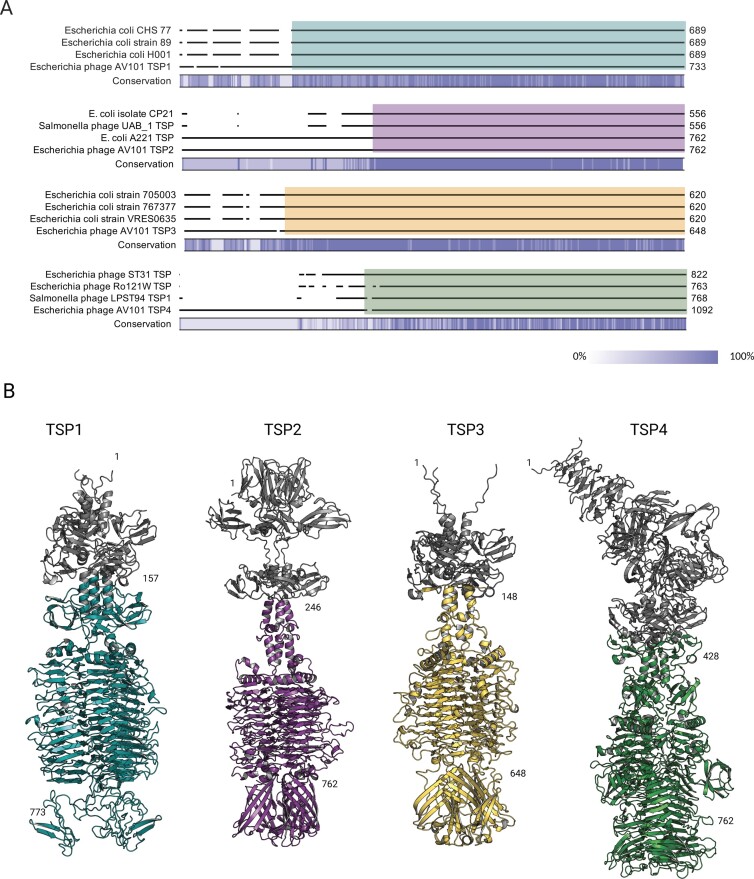
Figure 4.
The receptor-binding domain of AV101 TSPs show similarity to prophages and distant-related lytic phages. (A) BLASTp analysis of the four TSPs showed that TSP1, TSP2, and TSP3 share similarity towards proteins found in E. coli strains. Furthermore, TSP2 and TSP4 had similarity towards distant-related lytic phages. A detailed overview of the sequence similarity and the accession numbers of the BLASTp hits can be found in Table S3 (Supporting Information). The coloured boxes represent the region of similarity visualized in the structures. (B) AlphaFold 2 was used to prediction and visualize the four TSPs of AV101. The four TSPs folds like other known TSPs with the conserved β-helix serving as the receptor-binding domain. Moreover, the analysis showed that the amino acid sequence similarity to other TSPs were found in the receptor-binding domains, thus are highlighted in the same colours as in (A). The grey colour represents the N-termini structural domains; TSP1 amino acids 1–157; TSP2 amino acids 1–246; TSP3 amino acids 1–148, and TSP4: amino acids 1–428.