Acute aducanumab treatment triggers a concerted microglial response to amyloid, including increased peri-plaque recruitment and upregulation of activation, phagocytic, and immune-associated genes. However, discontinuation blunts microglial response up to 7 mo after the final treatment, even with the return of amyloid load.
Abstract
Aducanumab, an anti-amyloid immunotherapy for Alzheimer’s disease, efficiently reduces Aβ, though its plaque clearance mechanisms, long-term effects, and effects of discontinuation are not fully understood. We assessed the effect of aducanumab treatment and withdrawal on Aβ, neuritic dystrophy, astrocytes, and microglia in the APP/PS1 amyloid mouse model. We found that reductions in amyloid and neuritic dystrophy during acute treatment were accompanied by microglial and astrocytic activation, and microglial recruitment to plaques and adoption of an aducanumab-specific pro-phagocytic and pro-degradation transcriptomic signature, indicating a role for microglia in aducanumab-mediated Aβ clearance. Reductions in Aβ and dystrophy were sustained 15 but not 30 wk after discontinuation, and reaccumulation of plaques coincided with loss of the microglial aducanumab signature and failure of microglia to reactivate. This suggests that despite the initial benefit from treatment, microglia are unable to respond later to restrain plaque reaccumulation, making further studies on the effect of amyloid-directed immunotherapy withdrawal crucial for assessing long-term safety and efficacy.
Introduction
Aducanumab is a therapeutic monoclonal antibody that clears amyloid-β (Aβ) (Sevigny et al., 2016), a hallmark pathology of Alzheimer’s disease (AD), from the brain. It was the first Food and Drug Administration–approved disease-modifying treatment for AD, with many other anti-Aβ antibodies and Aβ-targeting therapies in development. Despite their popularity as a therapeutic strategy, the mechanisms behind Aβ clearance in many of these treatments, long-term effects, and effects of discontinuation have not been fully explored. Patients receiving these immunotherapies are in the early symptomatic stage of the disease and are assumed to require continuous treatment throughout the remainder of their lifetime to prevent further accumulation of Aβ and accompanying cognitive decline. However, anti-Aβ antibodies trigger an adverse vascular event known as amyloid-related imaging abnormalities (ARIA) (Sperling et al., 2011), often necessitating discontinuation. Financial constraints may also cause discontinuation due to the high cost of infusions. Therefore, understanding both (1) the long-term effects of aducanumab treatment and (2) the effects of cessation of treatment on plaques and other downstream contributors to disease is crucial to assessing the mechanisms underlying plaque clearance in aducanumab and other Aβ immunotherapies. Furthermore, understanding the basic biology driving the clearance of Aβ from the brain when aducanumab is administered allows for modulation of aspects of this mechanism to alternately enhance clearance and ameliorate side effects.
Microglia, the resident immune cells of the brain, play a critical role in the pathogenesis of AD (Song and Colonna, 2018; Leng and Edison, 2021). Although their role in disease progression is not fully understood, many AD risk genes (e.g., APOE and TREM2) are highly enriched in microglia. During AD, and in response to Aβ or surrounding neuronal damage, microglia transition from a ramified, homeostatic state to an amoeboid, phagocytic state is marked by the upregulation of various markers such as Trem2, ApoE, Cst7, Clec7a, Itgax, etc. (Keren-Shaul et al., 2017; Krasemann et al., 2017; Kang et al., 2018; Rangaraju et al., 2018; Sebastian Monasor et al., 2020). This state has been alternatively called DAM (disease-associated microglia) (Keren-Shaul et al., 2017), microglia in neurodegeneration (Krasemann et al., 2017), or simply “activated microglia” (Leng and Edison, 2021; Colonna and Butovsky, 2017; Chen and Colonna, 2021). Whether activated microglia are harmful or helpful is highly context-dependent and varies with disease type and stage. Microglia phagocytose Aβ plaques and are believed to also contribute to Aβ clearance during anti-Aβ immunotherapy (Sevigny et al., 2016; Wilcock et al., 2001, 2003, 2004), though the precise transcriptional and phenotypic changes that occur both during aducanumab treatment and after, and how these relate to plaque load and associated damage (e.g., neuritic dystrophy), are not understood. Neuritic dystrophy is a pathological feature of AD in both humans and amyloid mouse models (Benzing et al., 1993; Gowrishankar et al., 2015; Dickson et al., 1990; Onorato et al., 1989). Dystrophic neurites are large, swollen neuronal processes that amass an abnormal amount of cytoskeletal, lysosomal, and endosomal proteins and are closely associated with plaques (Gowrishankar et al., 2015; Nixon, 2007). In addition to measuring Aβ load directly, neuritic dystrophy can be measured often through markers such as LAMP1 (Gowrishankar et al., 2015; Cummings et al., 1992), APP (Cummings et al., 1992), or silver staining (Braak et al., 1989) as a readout of local neuronal damage.
To understand both the acute and chronic effects of aducanumab administration, and later withdrawal, on plaques, neuritic dystrophy, and microglial function and phenotype, we utilized the established treatment paradigm of delivering chimeric mouse aducanumab (henceforth referred to as “aducanumab”) that bears the humanized antibody’s variable region but with a mouse fragment crystallizable (Fc) region by intraperitoneal (IP) injection (Sevigny et al., 2016; Xiong et al., 2021; Da Mesquita et al., 2021) to APP/PS1 mice, a model of AD amyloidosis. We comprehensively assessed plaques, neuritic dystrophy, astrocytes, and microglia in both the acute treatment phase, when plaques were actively cleared, and during a protracted 7.5-mo washout phase, when treatment was ceased and plaques reaccumulated. We found previously unreported effects of aducanumab on plaque load as well as astrocyte and microglial dynamics, both acutely and during the long washout phase.
Results
Acute aducanumab treatment preferentially clears diffuse Aβ over fibrillar plaques while reducing neuritic dystrophy
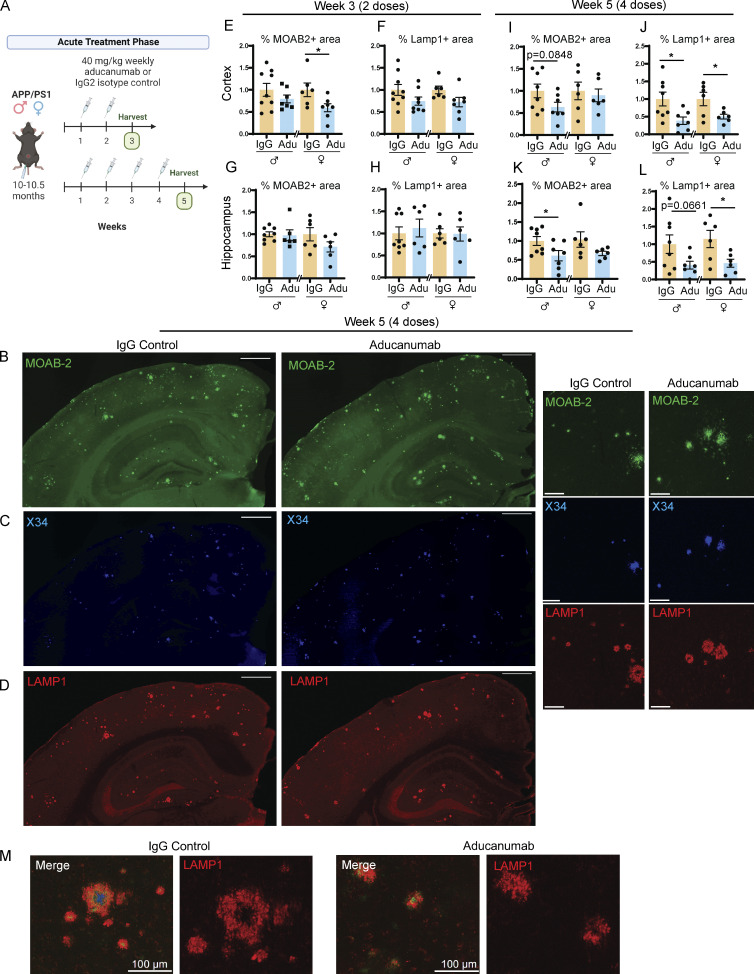
To investigate the acute effects of aducanumab treatment, we administered either two or four 40-mg/kg doses of aducanumab or immunoglobulin G (IgG) control by IP injection to 10-mo-old male and female APP/PS1 mice and harvested mice 1 wk after the final dose (Fig. 1 A). Using histology, we assessed total Aβ plaque load with the pan-Aβ antibody MOAB-2 (Fig. 1 B), fibrillar plaque load with the Congo red derivative X34 (Fig. 1 C), and neuritic dystrophy with LAMP1 (Fig. 1, D and M).
Figure 1.
Aducanumab modestly reduces Aβ and neuritic dystrophy in APP/PS1 mice during acute treatment phases. (A) Timeline of the acute treatment phase of the study. Male and female 10- to 10.5-mo-old APP/PS1 mice received either two or four doses of aducanumab or IgG control (40 mg/kg, IP injection) and were harvested for analysis 1 wk after the final dose. (B–D) Representative images of (B) total Aβ marked by MOAB-2, (C) X34+ fibrillar plaques, and (D) neuritic dystrophy marked by LAMP1, in male mice treated with four doses of aducanumab (Adu) or IgG control. (E and F) Quantification of percent of the tissue covered by MOAB-2+ (E) and LAMP1+ staining in the cortex (F), following two doses of aducanumab. (G and H) Quantification of percent of the tissue covered by MOAB-2+ (G) and LAMP1+ staining in the hippocampus (H), following two doses of aducanumab. (I and J) Quantification of the percent of tissue covered by MOAB-2+ (I) and LAMP1 staining in the cortex (J), following four doses of aducanumab. (K and L) Quantification of percent of tissue covered by MOAB-2+ (K) and LAMP1 staining in the hippocampus (L), following four doses of aducanumab. (M) LAMP1+ neuritic dystrophy surrounding amyloid plaques in IgG control and aducanumab-treated mice. Scale bars in B–D = 500 µm for whole tissue view, 100 µm for insets; scale bars in M = 100 µm. Data in E–L expressed as fold change relative to IgG control of the same sex. *P < 0.05; **P < 0.01, Student’s t test for normally distributed samples with no significant difference in variance (E; F; G females; H females; I; J males; K males; L females), Welch’s t test for normally distributed samples with differences in variances (J females; K females; L males), Mann–Whitney test for non-normally distributed samples (G males; H males), with males and females analyzed separately. N = 6–9 mice/sex/treatment.
Reduction of Aβ in aducanumab-treated versus IgG control mice was variable during this acute phase, similar to previous acute treatment studies (Xiong et al., 2021; Da Mesquita et al., 2021), though we detected modest reductions in total MOAB-2+ amyloid load in mice that received two or four doses of aducanumab (Fig. 1, E, I, and K). Changes in X34+ fibrillar plaque load between control and aducanumab-treated mice were even less pronounced (Fig. S1, A, I, Q, and Y), and average plaque size was larger in aducanumab-treated mice (Fig. S1, F and N), suggesting aducanumab more efficiently clears small, diffuse plaques over large, fibrillar plaques. The similarity in plaque clearance between the two and four dose groups suggests that, at least in the short-term, additional doses do not yield additional Aβ reduction.
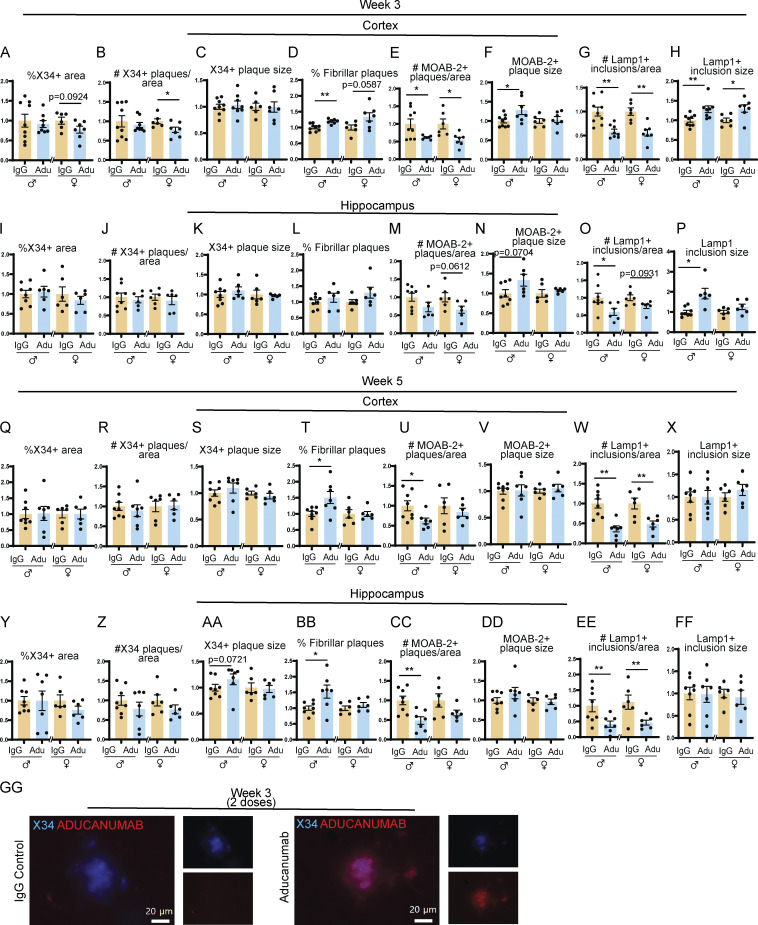
Figure S1.
Aducanumab modestly reduces total Aβ after two and four doses. (A–H) Quantification of plaques and neuritic dystrophy in the cortex after two weekly doses of IgG or aducanumab (Adu). (A) Percent of the tissue covered by X34+ staining (A), number of X34+ plaques per area (B), average X34+ plaque size (C), percent of X34+ fibrillar plaques (D), number of MOAB-2+ plaques per area (E), average MOAB-2+ plaque size (F), number of LAMP1+ inclusions per area (G), average LAMP1+ inclusion size (H). (I–P) The same as A–H, but in the hippocampus, after two weekly doses of IgG or aducanumab. (Q–X) The same as A–H, but in the cortex, after four weekly doses of IgG or aducanumab. (Y–FF) The same as A–H, but in the hippocampus, after four weekly doses of IgG or aducanumab. (GG) Staining of X34+ plaques with Cy3 mouse IgG secondary antibody to detect aducanumab in the acute treatment phase. *P < 0.05; **P < 0.01, Student’s t test for normally distributed samples with no significant difference in variance (A; B females; C; D; E females; F; G; H females; I; J males; K males; L; M; N males; O males; P females; Q; R males; S females; T females; U–Z; AA females; BB females; CC; DD; EE males; FF), Welch’s t test for normally distributed samples with differences in variances (E males; K females; N females; P males; BB males; EE females), Mann–Whitney test for non-normally distributed samples (B males; H males; J females; O females; R females; S males; T males; AA males), with males and females analyzed separately. N = 6–9 mice/sex/treatment.
To understand the downstream consequences of plaque clearance, we measured levels of LAMP1, an established marker of neuritic dystrophy that is used routinely in AD studies (Gowrishankar et al., 2015; Cummings et al., 1992; Barrachina et al., 2006; Hassiotis et al., 2018), including of murine aducanumab (Da Mesquita et al., 2021). Interestingly, despite the weak effect on plaque load itself, aducanumab strongly reduced neuritic dystrophy (Fig. 1, J and L) after four doses, suggesting that the drug could have additional therapeutic benefits outside the clearance of Aβ itself.
Microglial activation increases early in aducanumab treatment
Because microglia play a key role in AD, become highly activated in the presence of Aβ (Keren-Shaul et al., 2017; Kang et al., 2018; Krasemann et al., 2017), and are capable of phagocytosing plaques, we hypothesized that treatment with aducanumab further elevates microglial activation and engages pro-phagocytic microglial functions to aid in plaque clearance.
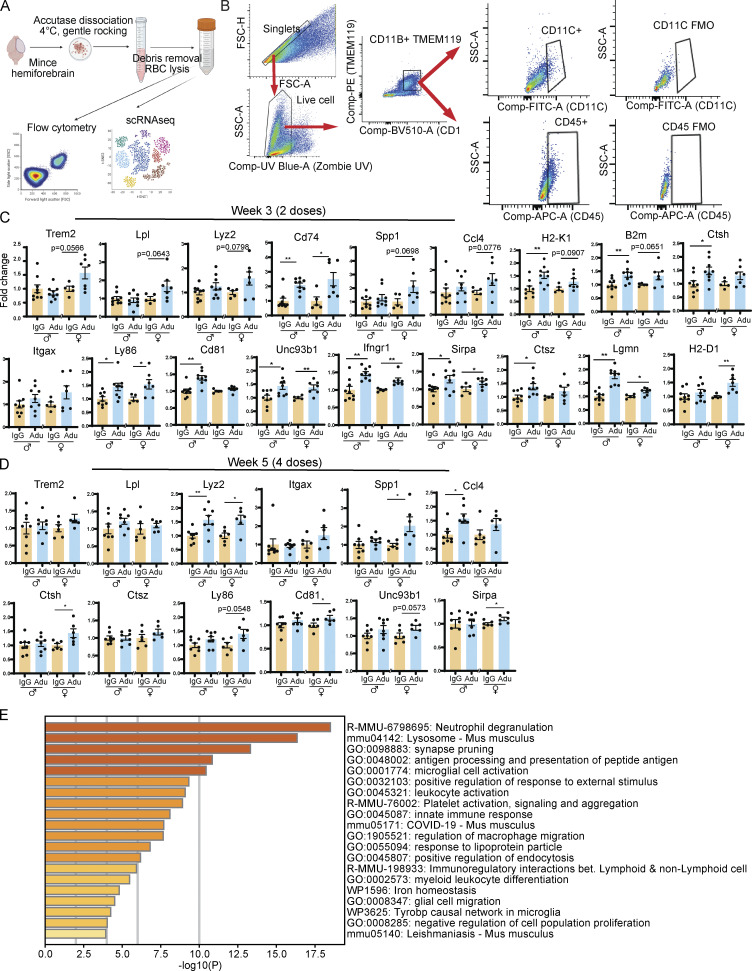
To assess the acute effect of treatment on microglia, we performed flow cytometry to measure the number of activated microglia following aducanumab treatment and RT-qPCR (reverse transcription-quantitative polymerase chain reaction) of RNA from bulk brain tissue to measure the expression of microglial activation markers. For flow cytometry, we defined microglia as cells that were both CD11b+ and TMEM119+, and within this population, activated microglia as cells that were additionally CD11c+ or CD45+ (Fig. S2 B). We observed an increase in the proportion of CD11c+ (Fig. 2 A) and CD45+ (Fig. 2 C) microglia that trended toward significance, and modest but significant increases in mean fluorescence intensity of CD11c (Fig. 2 B) in the CD11c+ microglia population, and CD45 (Fig. 2 D) in the CD45+ microglia population in aducanumab-treated mice after four doses. Accompanying these were increases in expression of microglial activation genes such as Lyz2, Cd74, Ccl4, and H2-D1 (Fig. 2 I and Fig. S2, C and D) as measured through RT-qPCR of bulk tissue. Interestingly, we observed the most dramatic increases in expression not of “classical” or “DAM-like” activation genes such as Trem2 or Lpl (Fig. S2, C and D), but of “MHC-like” activation genes, such as Cd74 and H2-D1 (Fig. 2 I and Fig. S2, C and D), which are involved in antigen presentation and processing. This suggests that while aducanumab does intensify existing DAM activation-like signatures present in the amyloid disease model, it may also confer microglia with a transcriptomic signature that is aducanumab-specific, prompting us to further explore this possibility with bulk and single-cell RNA sequencing (scRNAseq).
Figure S2.
Aducanumab increases microglial activation acutely. (A) Diagram showing dissociation methods used for flow cytometry and scRNAseq. (B) Flow cytometry gating strategy used for microglial analysis. Singlet gating axes—FSC-A: 0, 50K, 100K, 150K, 200K, 250K; FSC-H: 0, 50K, 100K, 150K, 200K. Live cell gating axes—UV Blue Zombie UV: −103, 0, 103, 104; SSC-A: 50K, 100K, 150K, 200K. CD11B+TMEM119+ gating axes—BV510 CD11B: −103, 0, 103, 104; PE TMEM119: −103, 0, 103, 104. CD11C+ gating axes—FITC CD11C: −103, 0, 103; SSC-A: 0, 50K, 100K. CD45+ gating axes—APC CD45: −103, 0, 103; SSC-A: 0, 50K, 100K. (C) RT-qPCR relative expression of microglial activation genes after two doses of aducanumab (Adu). (D) The same as C, but after four doses of aducanumab. All data in this figure expressed as fold change relative to IgG control within a single sex. (E) Metascape gene ontology heatmap generated from all genes significantly (P < 0.05) upregulated in aducanumab-treated mice. *P < 0.05; **P < 0.01, Student’s t test for normally distributed samples with no significant difference in variance (C: Trem2, Lpl males, Lyz2 males, Spp1 females, H2-K1, B2m males, Ctsh, Ly86 males, Cd81 females, Unc93b1 males, Ifngr1 males, Sirpa, Ctsz males, Lgmn, H2-D1 males; D: Trem2 males, Lpl, Lyz2, Itgax females, Spp1 males, Ctsh, Ctsz, Ly86, Cd81, Unc93b1, Sirpa), Welch’s t test for normally distributed samples with differences in variances (C: Lpl females, Lyz2 females, Ccl4 females, B2m females, Itgax females, Ly86 females, Unc93b1 females, Ctsz females; D: Spp1 females), Mann–Whitney test for non-normally distributed samples (C: Cd74, Spp1 males, Ccl4 males, Itgax males, Cd81 males, Ifngr1 females, H2-D1 females; D: Trem2 females, Itgax males, Ccl4), with males and females analyzed separately. N = 6–11 mice/sex/treatment.
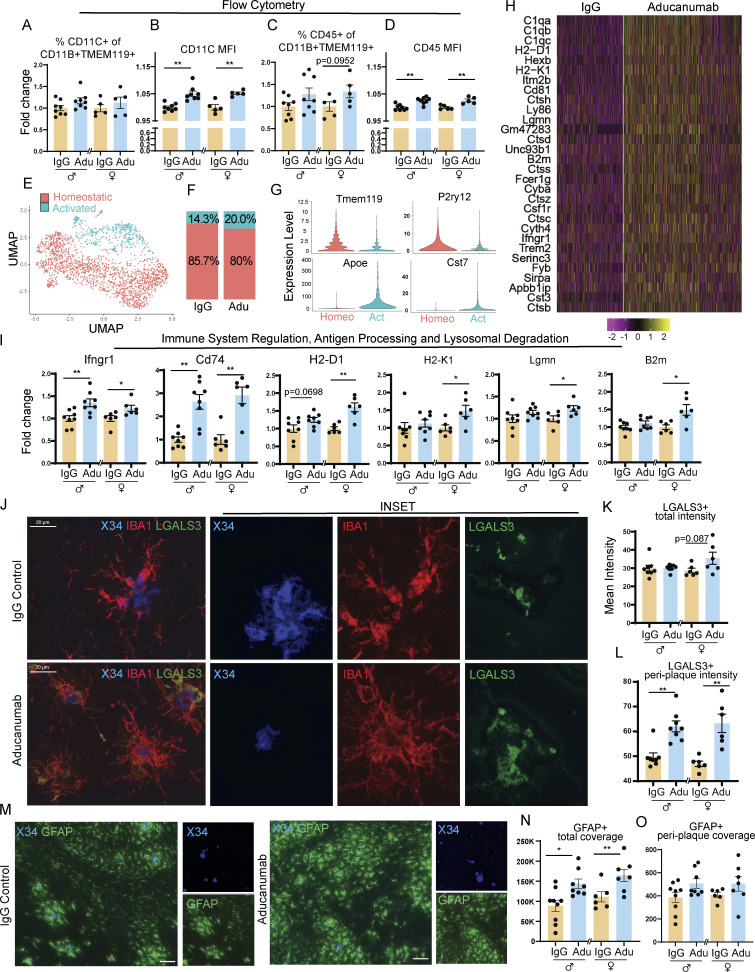
Figure 2.
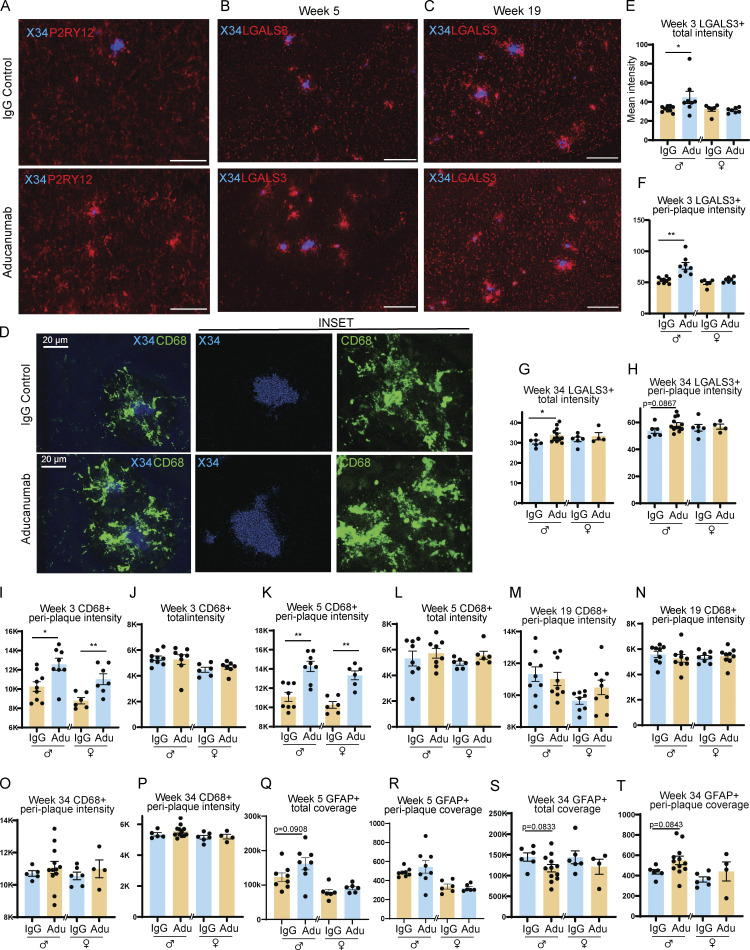
Aducanumab increases microglial activation, upregulates pro-clearance genes, and recruits microglia and astrocytes to plaques during acute treatment phases. (A–D) Flow cytometry data showing (A) percent of activated CD11C+ microglia, (B) mean fluorescence intensity of CD11C within the CD11C+ microglia population, (C) percent of activated CD45+ microglia, and (D) mean fluorescence intensity of CD45+ within the CD45+ microglia population, following four 40 mg/kg doses of aducanumab (Adu) or IgG control. Microglia were identified as CD11B+ TMEM119+ live cells. (E) UMAP projection of microglia subcluster from scRNAseq analysis, following four doses of aducanumab (N = 2 male mice/treatment). (F) Percent of homeostatic and activated cells in the microglia subcluster in aducanumab- or IgG-treated mice. (G) Violin plots of canonical homeostatic and activated microglial genes used to identify homeostatic and activated microglial subclusters. (H) Heatmap showing the top 30 most highly upregulated genes in aducanumab versus IgG control mice, with P_val_adj < 0.05. (I) RT-qPCR data from bulk hemiforebrain tissue showing relative expression of genes involved in immune system regulation, antigen processing, and lysosomal degradation, including validation of targets from the scRNAseq experiment. (J) Representative images of X34+ plaques and LGALS3+ and IBA1+ microglia following four doses of IgG control or aducanumab. (K) Quantification of total mean intensity of LGALS3+ microglia after four doses, related to J. (L) Quantification from Sholl analysis of mean intensity of LGALS3+ microglia 5 µm from plaques after four doses, related to J. (M) Representative images of X34+ plaques and GFAP+ astrocytes following two doses. (N) Quantification of total GFAP+ astrocyte coverage after two doses, related to M. (O) Quantification from Sholl analysis of GFAP+ astrocyte coverage 15 µm from plaques after two doses, related to L. Scale bars in J = 20 µm; scale bars in M = 100 µm. A–D and I expressed as fold change relative to IgG control of the same sex. *P < 0.05; **P < 0.01, Student’s t test for normally distributed samples with no significant difference in variance (A females; B females; C; D; I: Ifngr1, Lgmn, B2m, Cd74 males, H2-D1, H2-K1 females; K females; N), Welch’s t test for normally distributed samples with differences in variances (I: Cd74 females; L females; O females), Mann–Whitney test for non-normally distributed samples (A males; B males; I: H2-K1 males; K males; L males; O males), with males and females analyzed separately. N = 5–9 mice/sex/treatment.
scRNAseq reveals an aducanumab-specific microglial signature associated with antigen processing, lysosomal degradation, and immune system regulation
To further explore the aducanumab-specific microglial signature and to gain a higher-resolution picture of the molecular changes induced by aducanumab, we performed scRNAseq on the forebrain of male mice that received four doses of aducanumab or IgG control. We used the 10X Genomics platform and performed the analysis with Seurat V4.3 (Hao et al., 2021) (Fig. S2 A). After filtering, quality control, and cell population discovery, we subclustered microglia for further analysis (Fig. 2 E) and identified homeostatic and activated clusters based on the expression of canonical homeostatic (P2ry12, Tmem119) and activation (Apoe, Cst7) markers (Fig. 2 G).
The proportion of activated microglia was larger in aducanumab-treated mice (20.0%) compared with IgG-treated mice (14.3%) (Fig. 2 F), consistent with our flow cytometry data. Furthermore, we identified an aducanumab-specific microglial signature (Fig. 2 H) highly enriched for genes that are not only associated with classic microglial activation/DAM (C1qa, C1qb, C1qc, H2-D1, B2m, Ctsh, and Trem2) but are also highly involved in antigen processing and presentation (H2-D1, H2-K1, B2m, and Lgmn), lysosomal degradation (Ctsh, Ctsz, and Ctss), and other aspects of immune response and regulation, including cell migration (Ly86, Cd81, Unc93b1, Ifngr1, Fcer1g, and Sirpa). Indeed, gene ontology analysis revealed highly significant enrichment for processes involving the lysosome, phagosome, and endosome; microglial and innate immune cell activation; and regulation of macrophage and glial cell migration (Fig. S2 E). Many of these genes are also upregulated by microglia in disease models known to induce a pronounced inflammatory response, such as lipopolysaccharide and polyinosinic:polycytidylic acid injection (Kang et al., 2018). We validated the expression of these genes by RT-qPCR (Fig. 2 I). While several of these genes are associated with antigen processing and presentation, this signature is perhaps more reflective of an increased capacity for microglia to degrade phagocytosed material, such as Aβ, due to aducanumab treatment. Indeed, many of the genes most upregulated by aducanumab treatment, such as Lyz2, H2-D1, H2-K1, Ccl4, Cd74, and Spp1 are also highly expressed specifically by phagocytic microglia that have internalized amyloid (Grubman et al., 2021), suggesting the aducanumab treatment-specific signature is at least partly phagocytic.
Aducanumab increases the recruitment of activated microglia to plaques
Based on our data demonstrating that aducanumab increases microglial activation and confers microglia with a pro-phagocytic and pro-degradation gene signature, we posited that these changes in gene expression coincide with changes in microglial recruitment related to their enhanced ability to phagocytose and degrade amyloid plaques. To better understand the role of these activated microglia, we used histology to visualize microglia around plaques following treatment. After validating the presence of P2RY12+ (Fig. S5 A) and IBA1+ (Fig. 2 J) microglia around plaques in both aducanumab and IgG-treated mice, we additionally stained for LGALS3 (García-Revilla et al., 2022) to mark activated, plaque-associated microglia specifically (Fig. 2 J). Using Sholl analysis, we evaluated the intensity of LGALS3 staining 5 µm from plaques. At 3 wk, after two weekly doses, peri-plaque recruitment of LGALS3+ microglia was increased in male aducanumab-treated mice (Fig. S5 F), with an additional two doses yielding significant increases in microgliosis around plaques in both sexes (Fig. 2 L). We observed similar increased recruitment of CD68+ microglia in aducanumab-treated mice (Fig. S5, I and K), suggesting this effect is not specific to only a single gene or protein, but to activated microglia generally. In contrast to peri-plaque LGALS3+ microgliosis, total microgliosis was only slightly increased (Fig. 2 H and Fig. S5 E), suggesting that the effect of aducanumab on microglial reactivity is predominantly local. The increased recruitment of activated microglia to plaques suggests that aducanumab may promote plaque clearance and reduction of neuritic dystrophy through microglial phagocytosis and subsequent degradation of Aβ.
Figure S5.
Microglial and astrocytic recruitment to plaques during acute and washout phase. (A) Representative image of staining of X34+ plaques and P2RY12+ microglia following two doses of IgG control or aducanumab. Scale bar = 50 µm. (B and C) Representative image of staining of X34+ plaques and LGALS3+ microglia during (B) acute treatment phase after four doses, and (C) washout phase, 15 wk after four doses. Scale bar = 100 µm. (D) Representative image of staining of activated CD68+ microglia around X34+ plaques following two doses of IgG or aducanumab. Scale bar = 20 µm. (E) Quantification of total mean intensity of LGALS3+ microglia after two doses. (F) Quantification from Sholl analysis of intensity of LGALS3+ microglia 5 µm from plaques after two doses. (G) Quantification of total mean intensity of LGALS3+ microglia at 30 wk washout. (H) Quantification from Sholl analysis of intensity of LGALS3+ microglia 5 µm from plaques at 30 wk washout. (I) Quantification of total mean intensity of CD68+ microglia after two doses. (J) Quantification from Sholl analysis of intensity of CD68+ microglia 5 µm from plaques after two doses. (K) Quantification of total mean intensity of CD68+ microglia after four doses. (L) Quantification from Sholl analysis of intensity of CD68+ microglia 5 µm from plaques after four doses. (M) Quantification of total mean intensity of CD68+ microglia at 15 wk washout. (N) Quantification from Sholl analysis of intensity of CD68+ microglia 5 µm from plaques at 15 wk washout. (O) Quantification of total mean intensity of CD68+ microglia at 30 wk washout. (P) Quantification from Sholl analysis of intensity of CD68+ microglia 5 µm from plaques at 30 wk washout. (Q) Quantification of total coverage of GFAP+ astrocytes after four doses. (R) Quantification from Sholl analysis of coverage of GFAP+ astrocytes 15 µm from plaques after four doses. (S) Quantification of total coverage of GFAP+ astrocytes at 30 wk washout. (T) Quantification from Sholl analysis of coverage of GFAP+ astrocytes 15 µm from plaques at 30 wk washout. *P < 0.05; **P < 0.01, Student’s t test for normally distributed samples with no significant difference in variance (F females; G and H; I females; J–M; N males; O; P females; Q; R females; S; T males), Welch’s t test for normally distributed samples with differences in variances (F males; N females; P males; R males; T females), Mann–Whitney test for non-normally distributed samples (E; I males), with males and females analyzed separately. N = 4–11 mice/sex/treatment.
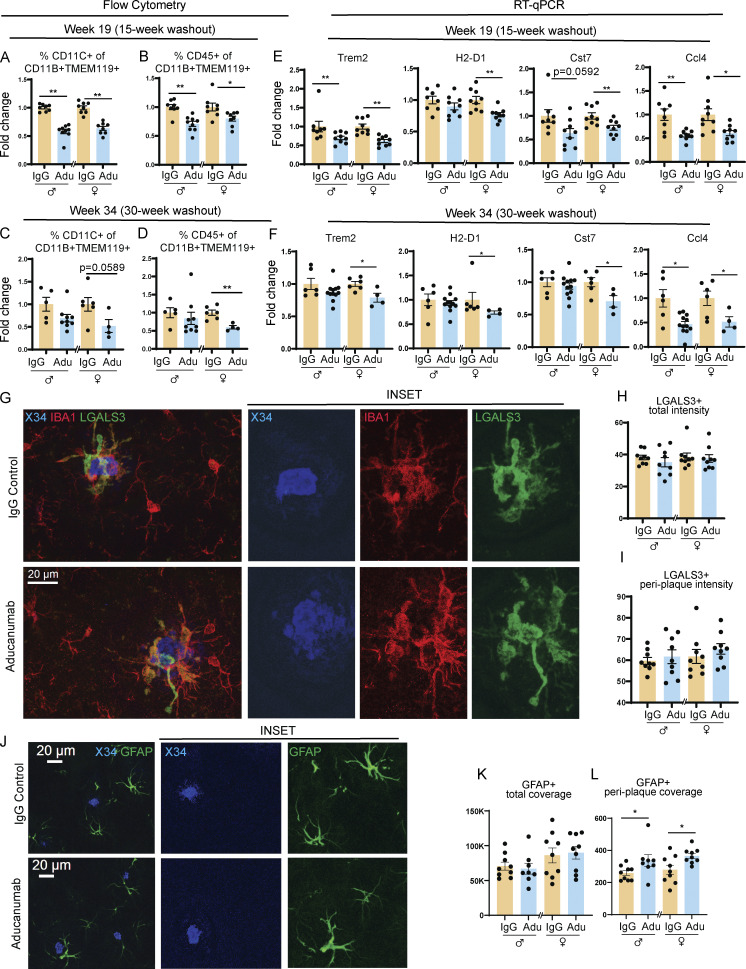
Because astrogliosis around plaques is also a feature of AD, we stained for glial fibrillary acidic protein (GFAP) at a dilution that predominantly detects only activated astrocytes (Fig. 2 M). After two doses, we observed an increase in total or overall astrogliosis in aducanumab-treated mice (Fig. 2 N) but only a trend toward increased peri-plaque astrogliosis (Fig. 2 O). This suggests that while acute aducanumab treatment increases astrogliosis, it does so in a global rather than local manner, unlike microglia, which exhibit more pronounced activation immediately adjacent to plaques. Whether different mechanisms drive global astrogliosis than local microgliosis and whether astrocytes perform similar or synergistic functions to microglia during plaque clearance remain open questions.
Global aducanumab-induced transcriptional changes are predominantly immune
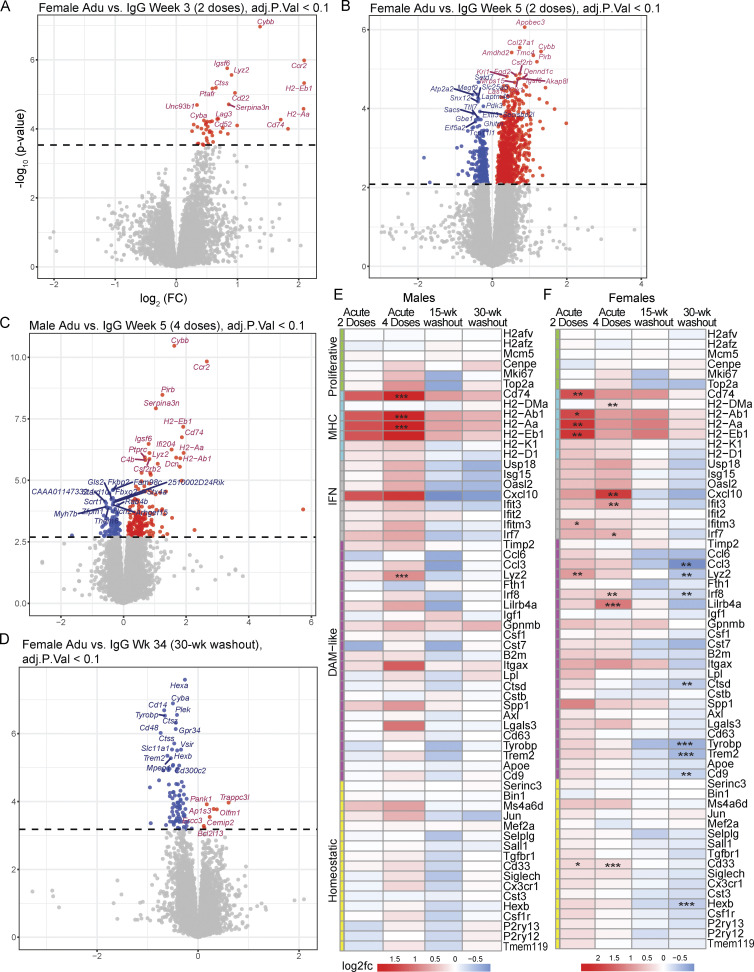
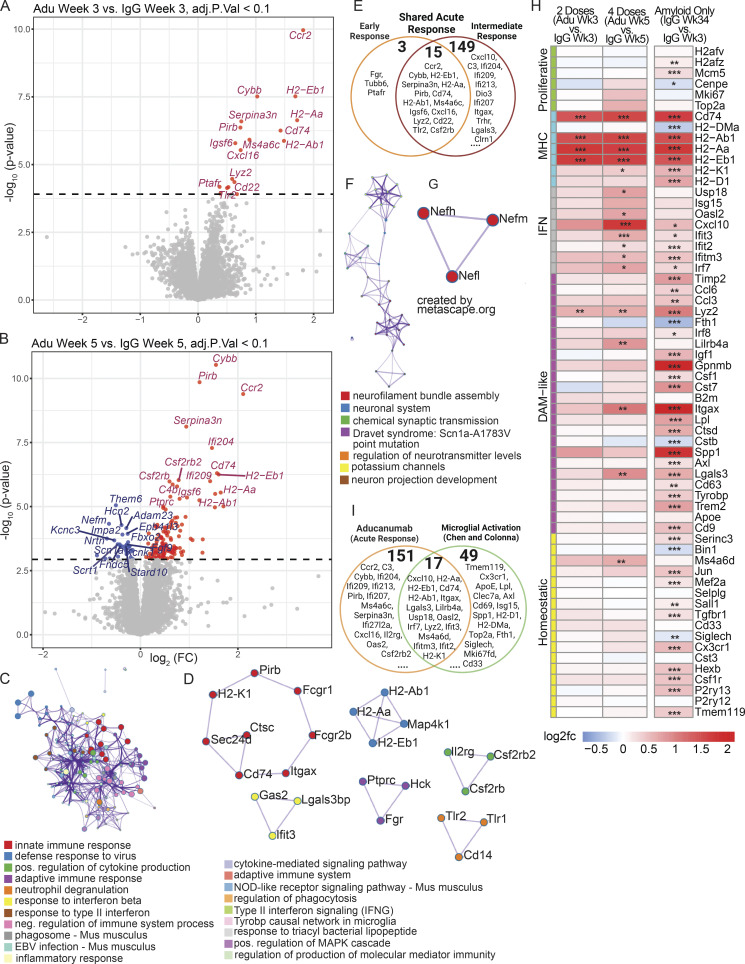
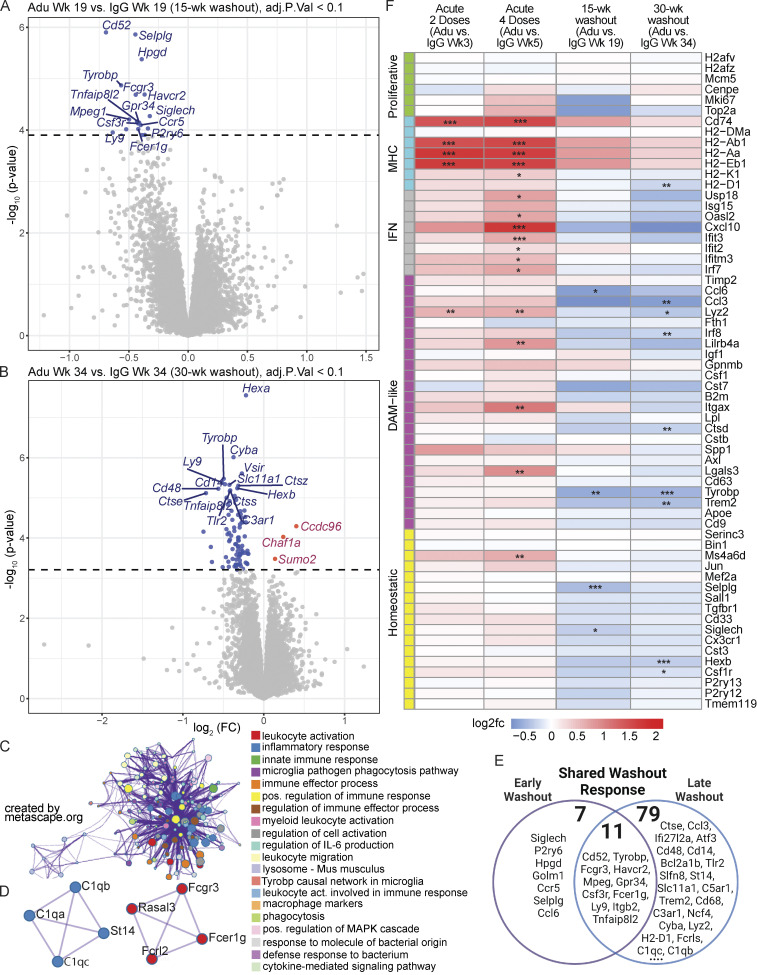
To further validate the aducanumab-induced transcriptional signatures we identified by scRNAseq and RT-qPCR, we performed bulk RNAseq on forebrain tissue from male and female aducanumab and IgG-treated mice at each study time point. This complementary independent approach provided additional depth and power for identifying subtler effects, including differences in response between sexes (Fig. S4). In agreement with our prior data, we identified a global acute aducanumab signature after two (Fig. 3 A) and four (Fig. 3 B) doses that consisted predominantly of immune and inflammatory genes such as Ccr2, Cybb, Pirb (Lilrb3), Serpina3n, Cd74, and Ifi204 (see Table S3 for full list). Indeed, gene ontology (Zhou et al., 2019) of transcripts upregulated during acute aducanumab treatment revealed highly significant enrichment for immune pathways, implicating interferon signaling, phagocytic regulation, and TYROBP causal networks (Fig. 3 C). Notably, TYROBP is an adaptor protein of TREM2, a gene that is strongly implicated in AD and is crucial for full DAM activation in mice. Similarly, protein–protein interaction networks (Fig. 3 D) predicted physical interactions between proteins belonging to multiple immune pathways, such as antigen processing and presentation (H2-Ab1, H2-Aa, H2-Eb1, and MAP4K1), JAK-STAT and cytokine-mediated signaling (IL2RG, CSF2RB2, CSF2RB), and adaptive immune response and Fc signaling (FCGR1, FCGR2B, ITGAX, CD74, CTSC, SEC24D, H2-K1, and PIRB). Though two doses of aducanumab were sufficient to induce activation, this effect was markedly magnified after four doses. To understand how additional doses impact the aducanumab signature, we compared differentially expressed genes (DEGs) between aducanumab and IgG groups at weeks three and five. Of the 167 genes differentially regulated after acute treatment, 15 were shared between the week 3 and 5 groups (“shared acute response”), with three genes uniquely expressed at week 3 (“early response”), and 149 genes uniquely expressed at week 5 (“intermediate response”) compared with respective IgG controls (Fig. 3 E). By stratifying our analysis by sex, we found that females (Fig. S4, A and B) responded earlier and more strongly to aducanumab treatment than males (Fig. S4 C), possibly as a result of the enrichment of innate immune genes on the X chromosome.
Figure S4.
Females respond earlier and more strongly to aducanumab, and display greater immune deactivation at washout. (A and D) Volcano plots showing DEGs from bulk RNAseq of bulk hemiforebrain tissue in (A) females after two doses of 40 mg/kg aducanumab (Adu) or IgG control (week 3), (B) females after four doses (week 5), (C) males after four doses (week 5), (D) females after 30-wk washout period following four doses (week 34). Gene names displayed are the top 15 upregulated and downregulated genes. (E–F) Heatmap of selected canonical microglial activation genes curated by Chen and Colonna (2021) from multiple studies of microglial activation, showing differential expression of these genes after two doses of aducanumab (week 3 aducanumab versus week 3 IgG), four doses of aducanumab (week 5 aducanumab versus week 5 IgG), 15-wk washout (week 19 aducanumab versus week 19 IgG), and 30-wk washout (week 34 aducanumab versus week 34 IgG) in (E) males and (F) females. N = 3–4 mice/sex/treatment/time point, significance at adjusted P val < 0.1 with FDR correction (statistics computed automatically with analysis in edgeR; see Materials and methods for additional details). For heatmaps, *P_val_adj < 0.1, **P_val_adj < 0.05; ***P_val_adj < 0.01.
Figure 3.
Bulk RNAseq reveals the global acute aducanumab signature is dominated by immune genes. (A and B) Volcano plots showing DEGs from bulk RNAseq of bulk hemiforebrain tissue following (A) two or (B) four 40 mg/kg doses of aducanumab (Adu) or IgG control. Gene names displayed are the top 15 upregulated and downregulated genes. (C and D) Metascape (C) gene ontology and (D) protein–protein interaction analysis of all genes upregulated in aducanumab treatment at weeks 3 and 5. (E) Venn diagram showing genes differentially expressed at either week 3 only (early response), week 5 only (intermediate response), or at both weeks (shared acute response). (F and G) Metascape (F) gene ontology and (G) protein–protein interaction analysis of all genes downregulated in aducanumab treatment at week 5. (H) Heatmap of selected canonical microglial activation genes curated by Chen and Colonna (2021) from multiple studies of microglial activation, showing differential expression of these genes after two doses of aducanumab (week 3 aducanumab versus week 3 IgG), four doses of aducanumab (week 5 aducanumab versus week 5 IgG), and aged amyloid only (week 34 IgG versus week 3 IgG, or 31 wk of aging in APP/PS1 mice). (I) Venn diagram showing the overlap of genes differentially expressed during aducanumab treatment with genes from the Chen and Colonna list. N = 4 mice/sex/treatment/time point, significance at adjusted P val < 0.1 with FDR correction (statistics computed with analysis in edgeR; see Materials and methods for additional details). For heatmaps, *P_val_adj < 0.1, **P_val_adj < 0.05; ***P_val_adj < 0.01.
A second key difference between response at weeks 5 and 3 was the downregulation of multiple neuronal genes at week 5, including Nefm, Nefl, Nefh, Them6, Impa2, and Adam23 (Fig. 3 B). Notably, neurofilament genes Nefm, Nefl, and Nefh are increased in both amyloid mouse models (George et al., 2017) and AD patients (He et al., 2020), with NEFL in plasma and cerebrospinal fluid used as a biomarker for AD. Gene ontology and protein–protein interaction analysis of genes downregulated by aducanumab treatment showed enrichment of primarily neuronal pathways (Fig. 3, F and G). The downregulation of neuronal damage-associated genes supports our earlier finding of reduced LAMP1+ neuritic dystrophy following aducanumab treatment, further suggesting immunotherapy-mediated neuroprotection that is not fully dependent on plaque reduction itself.
To contextualize the transcriptional aducanumab response within established microglial biology in neurodegeneration, we compared the aducanumab signature to a consensus microglial activation signature curated by Chen and Colonna (2021) from multiple studies of neurodegenerative disease models, including amyloidosis, tauopathy, and amyotrophic lateral sclerosis (Keren-Shaul et al., 2017; Zhou et al., 2020; Krasemann et al., 2017; Sala Frigerio et al., 2019; Hammond et al., 2019; Mathys et al., 2017; Friedman et al., 2018; Wang et al., 2020; Ellwanger et al., 2021; Li et al., 2019). We adopted their nomenclature, classifying genes as “homeostatic,” “DAM-like,” “interferon,” “MHC,” and “proliferative” (called cyc-M/cycling in the original review), and plotted the expression of these genes following two doses (week 3 aducanumab versus week 3 IgG), four doses (week 5 aducanumab versus week 5 IgG), and during increased amyloid accumulation without antibody treatment (week 34 IgG versus week 3 IgG) (Fig. 3 H). Interestingly, the acute aducanumab signature did not fully mirror the amyloid signature—although conventional DAM genes are slightly upregulated by treatment, the aducanumab signature is primarily driven by increased expression of MHC and interferon genes. Notably, the upregulation of many of these MHC and interferon genes with only two doses of aducanumab is equal to or even greater than the upregulation from 31 wk (7.75 mo) of amyloid accumulation. Furthermore, a subset of aducanumab-induced genes including Ccr2, C3, Cybb, and multiple interferon genes (Ifi204, Ifi209, Ifi213, Ifi207) are not in the consensus list identified by Chen and Colonna (Fig. 3 I). This suggests immunotherapy induces a transcriptional response that is at least partially distinct from the basal microglial response to increased amyloid pathology and is skewed heavily toward antigen processing/presentation and interferon signaling.
Collectively, our data from the acute treatment phase suggest that aducanumab engages pro-clearance microglial functions, including increased recruitment to plaques and phagocytosis and improved antigen degradation and processing, and confers microglia with a treatment-specific immune signature. The increase in microgliosis around plaques at week 5 compared with week 3 corresponds to the increased expression of activation-related genes observed both by RT-qPCR and bulk RNAseq, suggesting changes in gene expression are dose-dependent and play a synergistic role with peri-plaque recruitment in the response of microglia to aducanumab.
Reductions in plaque load and neuritic dystrophy are sustained long after cessation of treatment
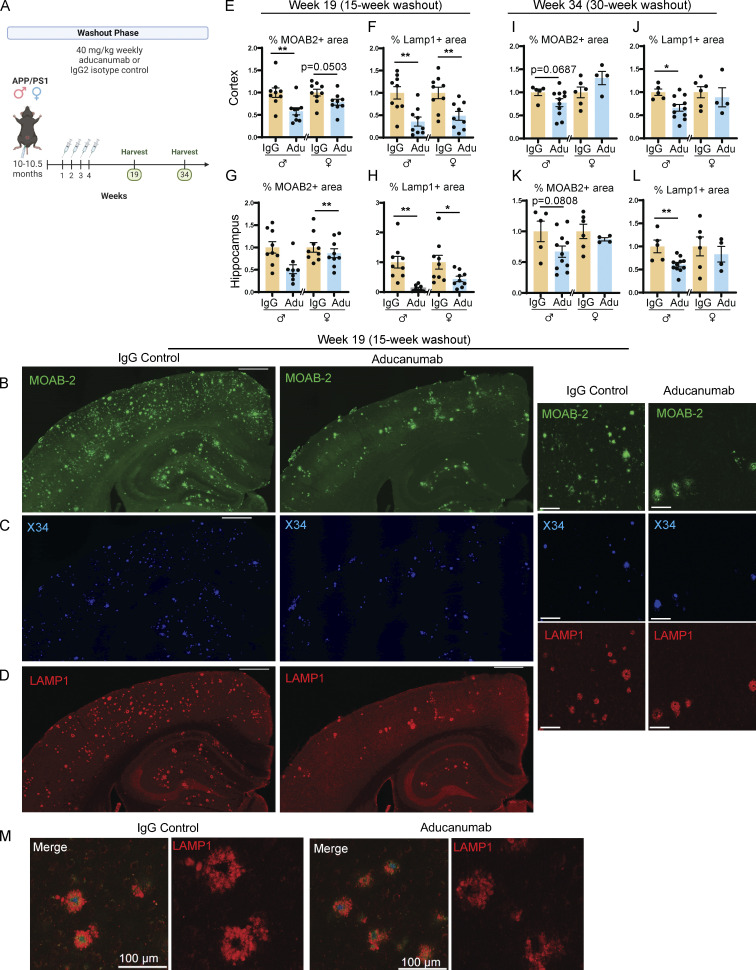
Not all patients who begin anti-Aβ immunotherapy will continue it throughout their lifetime, whether due to vascular side effects such as ARIA or financial constraints. Thus, we next assessed the impact of aducanumab discontinuation on plaque and microglial dynamics as this information is crucial to understanding the long-term effects of this or other anti-Aβ therapies. We again treated 10-mo-old APP/PS1 mice with four weekly doses of aducanumab or IgG control and harvested mice after a long washout period (either 15 or 30 wk later) to allow plaques to reaccumulate (Fig. 4 A). We analyzed total (Fig. 4 B) and X34+ fibrillar (Fig. 4 C) amyloid and associated neuritic dystrophy (Fig. 4, D and M).
Figure 4.
Reduction of amyloid load is sustained 15 but not 30 wk after cessation of aducanumab treatment. (A) Timeline of the withdrawal phase of the study. 10- to 10.5-mo-old male and female APP/PS1 mice received four doses of aducanumab or IgG control (40 mg/kg, IP injection) and were harvested either 15 or 30 wk after the final dose. (B–D) Representative images of (B) total Aβ marked by MOAB-2, (C) X34 fibrillar plaques, and (D) neuritic dystrophy marked by LAMP1, in male mice treated with four doses of aducanumab or IgG control, and harvested 15 wk after final treatment. (E and F) Quantification of the percent of the tissue covered by MOAB-2+ (E) and LAMP1+ staining in the cortex (F), following a 15-wk washout period. (G and H) Quantification of percent of the tissue covered by MOAB-2+ (G) and LAMP1+ staining in the hippocampus (H), following a 15-wk washout period. (I and J) Quantification of percent of tissue covered by MOAB-2+ (I) and LAMP1+ (J) staining in the cortex, following a 30-wk washout period. (K and L) Quantification of percent of tissue covered by MOAB-2+ (K) and LAMP1+ (L) staining in the hippocampus, following a 30-wk washout period. (M) LAMP1+ neuritic dystrophy surrounding amyloid plaques in following a 15-wk washout period. Scale bars in B–D = 500 µm for whole tissue view, 100 µm for insets; scale bars in M = 100 µm. Data in E–L expressed as fold change relative to IgG control of the same sex. *P < 0.05; **P < 0.01, Student’s t test for normally distributed samples with no significant difference in variance (E females; F; G; I females; J; K males; L), Welch’s t test for normally distributed samples with differences in variances (H; K females), Mann–Whitney test for non-normally distributed samples (E males; I males), with males and females analyzed separately. N = 4–11 mice/sex/treatment.
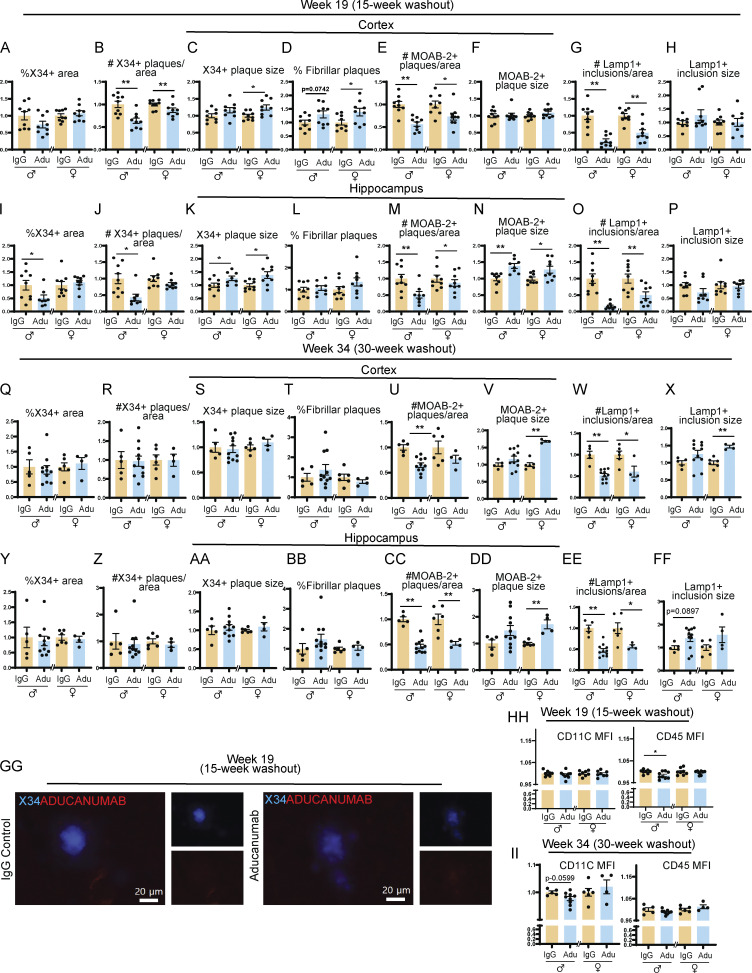
Surprisingly, even 15 wk after the final dose of aducanumab, plaque load not only remained low but was in fact lower than during the acute dosing phase. In both sexes, total Aβ plaque coverage (Fig. 4 E), number of X34+ fibrillar (Fig. S3 B), and total plaques (Fig. S3 E) were significantly reduced in the cortex. Similarly, the percent coverage of total Aβ was significantly reduced in females in the hippocampus, and while males also had reduced amyloid, this was not significant (Fig. 4 G). Neuritic dystrophy remained significantly lower in aducanumab-treated mice of both sexes and in both brain regions 15 wk after cessation (Fig. 4, F and H). Similar to what we observed in the acute treatment phase, the effect of aducanumab treatment on neuritic dystrophy was much stronger than its effect on plaques. These data suggest that not only is the effect of aducanumab sustained following cessation but that some of the mechanisms involved in clearing plaques and reducing dystrophy may even become more effective over time, even when the drug itself, which has a plasma elimination half-life of ∼21 days (Sevigny et al., 2016), is almost completely cleared from the system. Aducanumab, which colocalizes with plaques, is readily detectable in the brain by staining with anti-mouse IgG secondary antibody during the acute treatment phase (Fig. S1 GG), but is absent in the washout phase (Fig. S3 GG).
Figure S3.
Reduction in amyloid and neuritic dystrophy is sustained 15 but not 30 wk after aducanumab treatment. (A–H) Quantification of plaque and neuritic dystrophy in the cortex 15 wk after four weekly doses of IgG or aducanumab (Adu). Percent of the tissue covered by X34+ staining (A), number of X34+ plaques per area (B), average X34+ plaque size (C), percent of X34+ fibrillar plaques (D), number of MOAB-2+ plaques per area (E), average MOAB-2+ plaque size (F), number of LAMP1+ inclusions per area (G), average LAMP1+ inclusion size (H). (I–P) The same as A–H, but in the hippocampus, 15 wk after four weekly doses of IgG or aducanumab. (Q and X) The same as A–H, but in the cortex, 30 wk after four weekly doses of IgG or aducanumab. (Y–FF) The same as A–H, but in the hippocampus, 30 wk after four weekly doses of IgG or aducanumab. (GG) Staining of X34+ plaques and Cy3 mouse IgG secondary antibody to detect aducanumab in the washout phase. (HH) CD11C and CD45 mean fluorescence intensity from flow cytometry data from week 19. (II) CD11C and CD45 mean fluorescence intensity from flow cytometry data from week 34. Data in A–II expressed as fold change relative to IgG control within a single sex. *P < 0.05; **P < 0.01, Student’s t test for normally distributed samples with no significant difference in variance (A–D; E females; F–G; H females; I females; K–L; M males; N females; O females; Q females; R and S; T females; U–X; Y females; Z females; AA males; BB females; CC; DD males; EE males; FF females; HH: CD11C MFI, CD45 males; II), Welch’s t test for normally distributed samples with differences in variances (AA females; DD females; EE females), Mann–Whitney test for non-normally distributed samples (E males; H males; I males; J; M females; N males; O males; P; Q males; T males; Y males; Z males; BB males; FF males; HH: CD45 females), with males and females analyzed separately. N = 4–11 mice/sex/treatment.
In concordance with the decreased plaques and neuritic dystrophy at the 15-wk washout point, we observed a corresponding decrease in both the percent of CD11c+ (Fig. 5 A) and CD45+ activated microglia (Fig. 5 B) and the expression of activation markers including Trem2, Cst7, Ccl4, and H2-D1 (Fig. 5 E), indicating that microglia are in fact less activated following withdrawal of aducanumab treatment. Perhaps because Aβ burden remains low at this time point, microglia are able to deactivate in response to the blunting of this activation-inducing signal, demonstrating the plasticity of this activation phenotype.
Figure 5.
Microglial activation is blunted up to 30 wk after aducanumab treatment. (A and B) Flow cytometry data showing percent of activated (A) CD11C+ and (B) CD45+ microglia 15 wk after four doses of 40 mg/kg IgG control or aducanumab (Adu). (C and D) Percent of activated (C) CD11C+ and (D) CD45+ microglia 30 wk after four doses of 40 mg/kg IgG control or aducanumab. Microglia were identified as CD11B+ TMEM119+ live cells. (E and F) RT-qPCR from bulk hemiforebrain tissue showing relative expression of microglial activation genes (E) 15 wk or (F) 30 wk after four doses of IgG control or aducanumab. (G) Representative images of X34+ plaques and IBA1+ and LGALS3+ microglia 15 wk after four doses of IgG control or aducanumab. (H) Quantification of total mean intensity of LGALS3+ microglia 15 wk after four doses, related to G. (I) Quantification from Sholl analysis of mean intensity of LGALS3+ microglia 5 µm from plaques, 15 wk after four doses related to G. (J) Representative images of X34+ plaques and GFAP+ astrocytes 15 wk after four doses. (K) Quantification of total GFAP+ astrocyte coverage 15 wk after four doses, related to J. (L) Quantification from Sholl analysis of GFAP+ astrocyte coverage 15 µm from plaques 15 wk after four doses, related to J. Scale bars in G = 100 µm. Scale bars in J = 20 µm. A–F expressed as fold change relative to IgG control of the same sex. *P < 0.05; **P < 0.01, Student’s t test for normally distributed samples with no significant difference in variance (A females; B females; C females; D females; E: Trem2 females, H2-D1, Cst7 females; F: Trem2 females, H2-D1 males, Cst7, Ccl4 females; H males; I males; K; L females), Welch’s t test for normally distributed samples with differences in variances (E: Ccl4; F: Ccl4), Mann–Whitney test for non-normally distributed samples (A males; B males; C males; D males; E: Trem2 males, Cst7 males; F: Trem2 males; H females; I females; L males), with males and females analyzed separately. N = 4–11 mice/sex/treatment.
Microglia fail to fully reactivate after aducanumab withdrawal despite plaque recurrence
We assessed our final cohort of APP/PS1 mice 30 wk after the final of four aducanumab or IgG doses (Fig. 4 A). Although both amyloid plaque load and neuritic dystrophy burden were reduced at the 15-wk washout point (Fig. 4, E–H), plaque load in aducanumab-treated mice was similar to IgG controls by 30 wk (Fig. 4, I and K), though we still observed sustained reductions in neuritic dystrophy in male mice but not females (Fig. 4 J and Fig. 3 L). This suggests that the mechanism that restrains plaque reaccumulation at 15 wk is limited and eventually overcome by increasing plaque load, though it is notable that it takes between 15 and 30 wk—a comparatively long time for an aggressive amyloid overexpression mouse model like APP/PS1—for this to occur. As with the decoupled effect of aducanumab on plaques and neuritic dystrophy during the acute treatment phase, the sustained reduction of neuritic dystrophy in males 30 wk after withdrawal of treatment suggests aducanumab may exert a neuroprotective effect that is sustained even after plaque recurrence.
Intriguingly, microglia—which we hypothesized were deactivated at the 15-wk washout time point due to restrained plaque and neuritic dystrophy load—remained deactivated after 30 wk, despite the recurrence of plaques. The proportion of CD11c+ (Fig. 5 C) and CD45+ (Fig. 5 D) microglia in aducanumab-treated mice remained lower than in IgG-treated mice in females, and certain activation genes, including Trem2, Cst7, H2-D1, and Ccl4 (Fig. 5 F), remained downregulated. This implies that the microglial deactivation observed at the 30-wk washout point is not solely attributable to decreased amyloid but instead suggests an impairment in the ability of microglia to fully reactivate in the aftermath of treatment. This phenotype is not merely a return to baseline or pretreatment levels of microglial activation, as aducanumab-treated mice at week 34 (four doses of aducanumab followed by 30 wk of washout) display less activation than IgG control mice at the same time point (four doses of IgG followed by 30 wk of washout). Thus, mice that initially received aducanumab and then discontinued treatment displayed pronounced deactivation compared with mice of the same age that initially received IgG and then discontinued treatment. These sustained deactivation effects are stronger in females than males (Fig. 5, C, D, and F), and impaired microglial reactivation in females corresponds to reduced suppression of neuritic dystrophy and amyloid compared with males (Fig. 4, I–L).
In addition to the loss of the aducanumab treatment-specific signature, microglial recruitment to plaques (Fig. 5, G and I) and total microgliosis (Fig. 5 H) in the washout phase were no different from controls, suggesting the signals that drive increased microglial clustering around plaques is lost following withdrawal of the drug. However, the microglia that remain clustered around these plaques during the washout phase do not appear to be less activated in aducanumab-treated mice versus controls, suggesting aducanumab exerts disparate global versus local effects on microglia. Astrocytes also display differing local and global effects following aducanumab withdrawal (Fig. 5 J). Despite similar global astrogliosis between aducanumab and IgG-treated mice (Fig. 5 K), peri-plaque astrogliosis was in fact increased in aducanumab-treated mice during washout (Fig. 5 L). Aducanumab-induced peri-plaque astrogliosis appears only in the washout and not during acute treatment (Fig. 2 O), unlike microglia, that exhibit extensive peri-plaque microgliosis acutely (Fig. 2 L) but not during washout (Fig. 5 I). The exact causes of these treatment-specific disparate global and local effects on microgliosis and astrogliosis are unknown. When microglia are absent or microglial function is impaired, astrocytes are known to increase activation and phagocytosis as a compensatory mechanism (Konishi et al., 2020). Peri-plaque astrogliosis during washout may be a reaction to decreased microglial activation or reduced phagocytic capacity following withdrawal.
Immune pathways activated during acute aducanumab treatment are blunted following withdrawal
To further explore the microglial/immune deactivation signature, we again performed bulk RNAseq on forebrain tissue from males and females at both the 15-wk washout (Fig. 6 A) and 30-wk washout (Fig. 6 B). At both time points, we observed downregulation of predominantly immune genes (including Cd52, Tyrobp, and Fcgr3), pathways, and protein–protein interactions (Fig. 6, C and D), in agreement with our earlier findings of decreased microglial activation as measured by flow cytometry and RT-qPCR. The deactivation response was stronger at the late washout point than early washout point (Fig. 6 E), despite greater plaque recurrence at late washout, providing further support for the decoupling of immune activation and plaque load in the aftermath of aducanumab treatment.
Figure 6.
Immune pathways activated during acute aducanumab treatment are blunted following withdrawal. (A and B) Volcano plots showing DEGs from bulk RNAseq of bulk hemiforebrain tissue after a (A) 15- or (B) 30-wk washout period following four 40 mg/kg doses of aducanumab (Adu) or IgG control. Gene names displayed are the top 15 upregulated and downregulated genes. (C and D) Metascape (C) gene ontology and (D) protein–protein interaction analysis of all genes downregulated with aducanumab treatment at both washout points. (E) Venn diagram showing genes differentially expressed at either week 19 only (early washout), week 34 only (late washout), or at both weeks (shared washout response). (F) Heatmap of selected canonical microglial activation genes curated by Chen and Colonna (2021) from multiple studies of microglial activation, showing differential expression of these genes after two doses of aducanumab (week 3 aducanumab versus week 3 IgG), four doses of aducanumab (week 5 aducanumab versus week 5 IgG), 15-wk washout (week 19 aducanumab versus week 19 IgG), and 30-wk washout (week 34 aducanumab versus week 34 IgG). N = 3–4 mice/sex/treatment/time point, significance at adjusted P val < 0.1 with FDR correction (statistics computed automatically with analysis in edgeR; see Materials and methods for additional details). For heatmaps, *P_val_adj <0.1, **P_val_adj < 0.05; ***P_val_adj < 0.01.
Because the genes downregulated during washout and upregulated during acute treatment are both predominantly immune, we again plotted the expression of canonical microglial activation genes during acute treatment alongside washout (Fig. 6 F) to examine the relationship between aducanumab-induced acute activation and withdrawal deactivation. Strikingly, none of the activation genes induced acutely remained significantly elevated during washout. Instead, the washout signature is dominated by the downregulation of multiple DAM genes, including Trem2 and Tyrobp (see Table S6 for full list of DEGs), with pathway analysis again implicating the TYROBP microglial causal network and numerous other immune pathways (Fig. 6 C). Additionally, even among genes that are not significantly differentially regulated, nearly all modules, with the exception of the MHC module, displayed opposite aducanumab-mediated effects at washout than acutely. The MHC module is the only activation module that remained elevated during washout, suggesting that the blunting of microglial response in washout does not occur uniformly across all types of microglial activation genes. As with our acute data, where females responded more strongly to treatment, females also responded more strongly to withdrawal than males. When the analysis was stratified by sex, no DEGs were detected in males at either washout time point, while females downregulated many of the same genes at week 34 (Fig. S4 D) as in the combined analysis with both sexes (Fig. 6 B). Males displayed greater deactivation in the 15-wk washout period compared to the 30-wk washout period (Fig. S4 E), while females were more deactivated at the 30-wk washout period (Fig. S4 F). Thus, immunotherapy-responsive genes in males appear to respond more readily to increased plaque load than in females.
Collectively, the data from the washout phase demonstrate that microglia fail to fully reactivate many months after cessation of aducanumab treatment, despite—or perhaps even because of—early widespread activation. The aducanumab-specific microglia-immune signature observed acutely is not only lost but is reversed, demonstrating that withdrawal of treatment impacts microglia chronically.
Discussion
In this study, we sought to determine both the acute and long-term effects of aducanumab treatment on plaque and microglial dynamics in the APP/PS1 amyloid mouse model. We first demonstrated that acute aducanumab treatment induces widespread microglial activation at both the protein (Fig. 2, A–D) and RNA level (Fig. 2 I), increased microglial recruitment to plaques (Fig. 2, J and L; and Fig. S5), and adoption of a microglia-specific aducanumab gene signature enriched for immune, inflammatory, phagocytic, and endosomal/lysosomal genes (Fig. 2, E–H). We identified a bulk tissue-level aducanumab signature (Fig. 3, A and B) marked by upregulation of immune and inflammatory genes and pathways, including many that are microglia-specific (Fig. 3, C and D). While similar to known microglial signatures induced by amyloid alone, the aducanumab signature contained 151 novel genes (Fig. 3 I) and is dominated by interferon and MHC-related genes rather than DAM genes, suggesting the aducanumab-mediated immune signature is not fully captured by established microglial biology in AD. The upregulation of genes associated with phagocytosis, antigen processing, and protein degradation in particular (Fig. S2 D and Fig. 3, C and D) suggests that aducanumab activates a transcriptional network to enhance the breakdown of phagocytosed material, thus improving microglia-mediated plaque clearance. The implication of TYROBP signaling in these networks provides a possible starting point for further mechanistic investigation, as TREM2 signaling is important for achieving full DAM activation, and may also play a role in antibody-induced activation. Although bulk RNAseq does not provide the resolution required to identify the cellular origin of these transcripts, the enrichment for predominantly immune genes strongly suggests they are produced by immune, and likely myeloid cells. Furthermore, microglia from our scRNAseq data which were marked by expression of canonical myeloid genes such as P2ry12 and the microglia-specific Tmem119 (Fig. 2 G) showed similar enrichment for immune and inflammatory genes (Fig. 2 H). Though we cannot rule out the possibility of non-myeloid immune contributions to the bulk aducanumab signature, canonical T cell markers Cd8 and Cd4 were not differentially expressed in aducanumab-treated mice at any time point, suggesting these inflammatory responses are not caused by infiltrating T cells.
The exact microglial mechanisms that contribute to plaque clearance and neuritic dystrophy require further investigation. Increased microglial transcriptional activation and recruitment to plaques may work synergistically to promote amyloid clearance and reduce neuritic dystrophy. Increased microgliosis around plaques at week 5 (Fig. 2 L) compared with week 3 (Fig. S5 E) corresponded to increased expression of activation genes (Fig. 3, A, B, and I), suggesting these two readouts may be linked functionally. Aside from their ability to phagocytose amyloid, microglia also form a tight physical barrier around plaques, restricting the further spread of plaques and neuritic dystrophy (Condello et al., 2015). Ablation of microglia via CSF1R inhibition is known to increase neuritic dystrophy in the 5xFAD amyloid mouse model (Spangenberg et al., 2019); perhaps the role these cells play in restricting neuritic dystrophy basally is enhanced by aducanumab. Our transcriptomic data showing the reduction of neuronal genes, particularly neurofilament genes (Fig. 3, B, F, and G), provide further support for reductions in neuritic dystrophy observed acutely. Aducanumab is also known to increase phagocytosis of Aβ-42 by primary cultured microglia in vitro (Sevigny et al., 2016); our data support this finding in vivo.
In addition to identifying a treatment-specific acute aducanumab signature, we also demonstrated that microglial activation is strongly blunted by withdrawal of aducanumab up to 7.5 mo after the final dose. Microglia from mice treated with aducanumab do not merely return to baseline pretreatment levels of activation—they are less activated than their IgG control counterparts, implying that the treatment and subsequent withdrawal of aducanumab itself dampens later microglial response. While the acute aducanumab signature was marked primarily by the upregulation of interferon and MHC-like genes, the washout signature is marked by the downregulation of DAM genes (Fig. 6 F). This effect persisted even in the presence of recurring amyloid load, implying microglia may be unable to respond to later plaque accumulation.
Our findings from the acute and withdrawal phases of our study outline a biphasic effect of aducanumab on microglial activation and transcriptional profiles. Early in treatment, aducanumab engages a transcriptional network geared toward enhanced microglial phagocytosis and degradation of phagocytosed material, increases proportions of these activated microglia, and improves recruitment to plaques. These effects all likely synergize to improve plaque clearance. However, at later stages following treatment discontinuation, this phenotype reverses and microglia remain locked in a deactivated state, even as plaque load reaccumulates 7.5 mo later. The precise cause of this failure to reactivate is unclear but may be an “exhaustion-like” response caused by the initial highly activated state induced by aducanumab 30 wk earlier. This interpretation is supported by previous data from other active and passive immunization studies, showing elevated microglial activation after five, but not nine Aβ inoculations, that authors attributed to desensitization of microglia to anti-Aβ antibodies (Wilcock et al., 2001), and peaks in activation from passive immunization after 1–2 mo of treatment, followed by a return to baseline at 3 mo (Wilcock et al., 2004). Thus, although microglial activation may be beneficial in clearing plaques at early and intermediate time points, excessive activation may result in an impaired ability to reactivate later. Though we have not assessed microglia in a withdrawal period beyond 30 wk (7.5 mo), the failure of these cells to fully reactivate even at this protracted time point suggests the loss of one of the brain’s key mechanisms for restraining plaque load. We and others previously observed that microglial activation is increased in untreated female versus male APP/PS1 mice of the same age (Kang et al., 2018), and data from our current study indicate stronger activation in females (Fig. S4). Perhaps microglia from females, which are more activated basally in amyloid models, are also more readily activated by aducanumab during acute treatment, and thus more prone to deactivation after withdrawal.
Our data also help reconcile findings from two previous aducanumab studies in the 5xFAD model, showing either decreased expression of activation genes after eight weekly doses of aducanumab (Da Mesquita et al., 2021) or increased activation after four doses administered across 10 days (Xiong et al., 2021). In both studies, mice were analyzed shortly after the final dose. These seemingly contradictory results are clarified by the data from our dual paradigm study that show microglial activation is increased acutely, shortly after the first few doses, but that subsequent chronically administered doses may exhaust these cells, rendering them unable to reactivate. Thus, the response of microglia seems to depend not only on the amount in an individual dose or the number of doses but also on the duration of time between doses, and the length of the discontinuation period. The failure of microglia to reactivate after withdrawal may be problematic for patients who discontinue the drug. Biogen, the manufacturer of aducanumab, has reported that Aβ load remains low a year after the final dose in a subset of patients followed after termination of clinical trials. However, microgliosis is not easily assessed in living patients, and longer-term effects remain unclear. Further studies on the effects of chronic dosing and withdrawal are necessary as these therapies gain wider adoption. An adjunct treatment targeting microglia specifically may help enhance the effect of aducanumab by increasing microglial health and function, both during and after treatment.
Materials and methods
Study design
This study aimed to assess the effect of aducanumab treatment and withdrawal on plaque and microglial dynamics. To assess the effect of acute treatment, we administered either two or four weekly doses of IP aducanumab or IgG control to male and female 10- to 10.5-mo-old amyloid APP/PS1 mice, an age when these mice develop moderate plaque loads. We then harvested these mice 1 wk after the final dose. To assess the effect of withdrawal of treatment, we administered four weekly doses of aducanumab or IgG control to male and female 10- to 10.5-mo-old APP/PS1 mice and harvested mice either 15 or 30 wk after the final dose. We performed RT-qPCR, flow cytometry, and staining/histology to assess plaque load, neuritic dystrophy, and microglial activation, respectively. Each of these assays was performed on N = 5–12 mice/sex/treatment. We also performed scRNAseq on a subset (N = 2/treatment) of male mice in the acute group that received four doses and bulk RNAseq on N = 4 mice/sex/treatment/time point, except for the week 34 aducanumab female group (N = 3). Mouse numbers are detailed in figure legends and graphs are plotted to show individual data points.
Animals
Male and female APPswe/PS1ΔE9 (APP/PS1) mice (10–10.5 mo) on a pure C57BL/6J background were used. All animals had been backcrossed to C57BL/6J mice for at least 10 generations. All animals used in the study were bred in-house. Animals were housed under standard conditions with ad libitum access to food and water in a specific pathogen–free environment at Mayo Clinic Arizona. All studies were done in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals under an approved protocol from the Mayo Clinic Institutional Animal Care and Use Committee.
Aducanumab administration
Aducanumab and IgG2a control were generated by Absolute Antibody through transfection of HEK293 cells followed by protein A purification. Aducanumab heavy and light chain variable regions were obtained from the original patent (WO 2016/087944) (Xiong et al., 2021).
Mice were administered either chimeric mouse aducanumab or IgG control via IP injection (40 mg antibody/kg mouse) once a week for 4 wk. Mice were harvested at the following time points: week 3 (1 wk after the second injection), week 5 (1 wk after the fourth injection), week 19 (15 wk after the fourth injection), and week 34 (30 wk after the fourth injection).
Brain harvest and preliminary tissue processing
Mice were deeply anesthetized with Euthasol and intracardially perfused with 25 ml of ice-cold PBS with 0.2% 0.5 M EDTA. One hemiforebrain was fixed in 4% paraformaldehyde at 4°C for 48 h, before being transferred to a 30% sucrose in PBS solution. The other hemiforebrain was quickly diced into very fine pieces with a razor with ∼30 mg of tissue snap frozen and reserved for qPCR and bulk RNAseq, while the remaining tissue was processed for flow cytometry and scRNAseq.
Accutase dissociation for flow cytometry and scRNAseq
The minced hemiforebrain was suspended in 10 ml ice-cold Accutase in PBS (07922; StemCell Technologies) and dissociated for 30 min at 4°C on a rocker. The cell suspension was centrifuged at 300 × g for 10 min at 4°C and the supernatant was removed. Cells were resuspended in 5 ml DPBS with calcium, magnesium, glucose, and pyruvate (14287080; Gibco). The solution was sequentially triturated 10 times each with 10 ml, 5 ml, and P1000 pipettes, and then filtered through a 70-μm strainer. The resulting cell suspension was centrifuged at 300 × g for 10 min at 4°C. The supernatant was aspirated, and myelin and debris were removed with the Miltenyi Debris Removal Kit according to the manufacturer’s instructions. Red blood cells were removed with Miltenyi Red Blood Cell Lysis solution according to the manufacturer’s instructions. The final single-cell suspension was divided into two aliquots for either flow cytometry or scRNAseq.
Flow cytometry
Cells were washed in 1 ml Cell Staining Buffer (420201; BioLegend) and centrifuged at 400 × g for 5 min at 4°C. The supernatant was removed and the cells were resuspended in 100 μl 1:500 Zombie UV viability dye in PBS (423108; BioLegend) and incubated in the dark for 15 min at room temperature. Following viability staining, 1 ml Cell Staining Buffer was added to each sample and samples were centrifuged at 400 × g for 5 min at 4°C. The supernatant was removed, and cells were blocked in 25 μl 1:25 anti-mouse CD16/32 antibody in Cell Staining Buffer (156604; BioLegend) on ice for 10 min in the dark. 25 μl of antibody solution was then added to each tube for a final concentration of 1:100 for each antibody: PE anti-TMEM119 (ab225496; Abcam), BV510 anti-CD11B (101245; BioLegend), FITC anti-CD11C (117305; BioLegend), and APC anti-CD45 (109813; BioLegend). Cells were stained on ice in the dark for 30 min. Cells were washed in 1 ml Cell Staining Buffer and centrifuged at 400 × g for 5 min at 4°C. Cells were resuspended in ∼300 μl of cell staining buffer. UltraComp eBeads Plus Compensation Beads (01-3333-41; Invitrogen) were used for compensation controls according to the manufacturer’s instructions, and fluorescence minus one (FMO) controls for each antibody were also used. Samples were run on a BD LSRFortessa at the Mayo Clinic Scottsdale Flow Cytometry Core. Analysis was performed in FlowJo v10.8.1. FMO controls were used to determine positive signal for each fluor.
scRNAseq gel bead in emulsion (GEM) generation, library preparation, and sequencing
Cells were counted and processed for GEM generation according to the manufacturer’s instructions (10X Genomics) using the v3.1 GEM generation and library preparation kits. Cells were loaded onto a Chromium Single Cell Chip G with reverse transcriptase master mix, gel beads, and partitioning oil. GEMs were generated on the 10X Chromium Controller. Libraries were generated according to the manufacturer’s instructions. Samples were sequenced on a NovaSeq S4 at the Mayo Clinic Rochester Genome Analysis Core.
scRNAseq analysis
Reads were aligned to the mm10 mouse reference genome, and then aggregated and normalized with Cell Ranger (10X Genomics). Count matrices were loaded into R and analyzed with Seurat V4. All samples were downsampled to 6,000 cells and merged into a single Seurat object. Low-quality cells were removed by filtering out cells with <200 and >5,000 features, >5% of reads originating from mitochondrial genes, and >10% of reads originating from ribosomal genes. A preliminary round of clustering was performed. Briefly, SCTransform was run to normalize and scale data and regress out percent.mt, percent.hb, and percent.ttr (respectively, percent of transcripts from mitochondrial genes, hemoglobin genes, and transthyretin, a gene highly expressed in choroid cells that often contaminate scRNAseq datasets [Olney et al., 2022]). Dimensionality reduction using the first 50 principal components was performed, followed by the construction of the shared nearest neighbor graph using 30 dimensions, Louvian clustering at resolution 0.4, and visualization of cell populations with UMAP using 30 dimensions. Cell populations were identified using canonical markers for neurons (percent.neuron: Snap25, Syt1, Gad1, Gad2), oligodendrocytes (percent.oligo: Plp1, Mbp), astrocytes (percent.astro: Clu, Gfap, Aqp4), microglia (percent.micro: Hexb, C1qa), oligodendrocyte precursor cells (percent.opc: Olig1, Pdgfra, Vcan), and endothelial cells (percent.endo: Rgs5, My19, Igfbp7, Fn1, Sox17). The microglia cluster was isolated for further cleaning and analysis. Scaling, normalization, and clustering were performed on microglia as previously described. Contamination from ambient RNA of other cell types was identified by visualizing the expression of percent.neuron, percent.oligo, percent.astro, percent.opc, and percent.endo on Violin and Feature Plots, and cells were filtered to retain only those with percent.oligo <1, percent.astro <0.15, and percent.neuron <0.5. A final round of scaling, normalization, and clustering was run, this time with a clustering resolution of 0.1, to yield the final cleaned microglia dataset. The homeostatic and activated microglia subclusters were identified based on the expression of canonical homeostatic (Tmem119, P2ry12) and activated (Lyz2, Cst7, Apoe, Trem2) markers, and a differential expression test with the FindMarkers function validated this categorization. Finally, aducanumab-specific microglial genes were identified in the entire microglial population using the FindMarkers function. A heatmap of the top 30 most upregulated genes with adjusted P value <0.05 was plotted with the DoHeatmap function using the scaled data as input. Raw data are deposited in the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA1026618), and code is available at https://github.com/fryerlab/aducanumab/tree/main/scRNA.
RT-qPCR
RNA was extracted from bulk frozen tissue with the RNeasy Plus Mini Kit (Qiagen) according to the manufacturer’s instructions. cDNA was generated from RNA using iScript Reverse Transcription Supermix (1708840; Bio-Rad) according to the manufacturer’s instructions. qPCR reactions were set up with iTaq Universal Sybr Green Supermix (Bio-Rad), 400 nM primer mix, and 100 ng cDNA template per 20 μl reaction. Amplification was performed on a Roche LightCycler480 System.
Bulk RNAseq
N = 4 samples/sex/treatment/time point were used for bulk RNAseq, except for the aducanumab week 34 female group (N = 3; one sample was excluded because the sex annotated in the metadata did not match genetic sex as measured by Xist expression, and this sample was also excluded from all other assays). Representative samples were selected based on qPCR data (i.e., the four samples in each group with the closest values to the group average). Library preparation and sequencing were performed by the Mayo Clinic Rochester Genome Analysis Core. All RNA had RNA integrity number ≥ 8.5 (average 9.06). DNase I double digests were performed prior to library preparation with the Illumina Stranded Total RNA Prep with Ribo-Zero Plus kit according to the manufacturer’s instructions. All samples were sequenced simultaneously across an entire Illumina NovaSeq 600 S4 PE100 flow cell at 100 × 2 paired-end reads for an average of 170 million read pairs.
Bulk RNAseq analysis
BBDuk, part of the BBMap package, was used to trim Nextera adapter sequences off of reads. The recommended adapter trimming settings from the BBDuk guide were used in addition to polyA and polyG trimming (Bushnell, 2014). FastQC (Andrews, 2010) and MultiQC (Ewels et al., 2016) were used to assess the quality of raw and trimmed reads. Trimmed reads were aligned to the mouse reference genome mm10 using STAR with default settings (Dobin et al., 2013). Gene features were counted using featureCounts, part of the subread package, and mm10 annotation. Only primary alignments were counted, and reads were reversely stranded (Liao et al., 2014). A count matrix was generated containing the gene count for each sample.
Downstream analysis was limited to protein-coding genes and Xist. Lowly expressed genes were removed; genes were kept if they expressed at least 1 count per million in at least four samples. Library sizes were converted into effective library sizes by calculating normalization factors using the Trimmed Mean of M-values (TMM) with the edgeR package (Robinson et al., 2010). The remaining genes and samples were used as input for the limma/voom pipeline. Counts were log-transformed and sample-specific and observational-level weights were applied using the limma package (Ritchie et al., 2015; Law et al., 2014). For analyses that included both male and female samples, a model was created to compare groups (distinguished by treatment and dose) and sex was added as a cofactor. For sex-specific analyses, a model was created to compare groups (distinguished by treatment, dose, and sex). Differential expression was performed between groups, and the P value was adjusted with the Benjamini-Hochberg false discovery rate. Genes were identified as differentially expressed at adjusted P < 0.1. Raw data are deposited in the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA1026618), and code is available at https://github.com/fryerlab/aducanumab/tree/main/bulkRNA. The list of DEGs is included in the online supplemental material (acute phase in Table S3, washout phase in Table S6, and sex-specific analysis in Table S10).
Metascape gene ontology
Metascape gene ontology analysis (Zhou et al., 2019) was performed using the “express analysis” setting. Figs. 3, 6, and S2 show gene ontology heatmaps, networks, and protein–protein interaction networks. For the scRNAseq experiments, the full list of significantly upregulated genes in aducanumab-treated mice from the scRNAseq experiment was used as input for. For bulk RNAseq (acute upregulated, Fig. 3, C and D), the full list of genes significantly upregulated by aducanumab acutely was used as input. For bulk RNAseq (acute downregulated, Fig. 3, F and G), the full list of genes significantly downregulated acutely was used as input. For bulk RNAseq (washout downregulated, Fig. 6, C and D), the full list of genes significantly downregulated during washout was used as input.
Immunofluorescence of mouse tissue
Hemiforebrains were frozen in optimal cutting temperature compound and cut coronally with a freezing sliding microtome to generate 50-μm sections. Sections were washed in PBS and then stored in cryoprotectant at −20°C until further use. Sections were mounted onto slides, washed three times with PBS, and then permeabilized with PBS-X (PBS with 0.3% Triton-X) at room temperature for 30 min. Sections were blocked with 1% non-fat dry milk in PBS-X at room temperature for 1 h. Primary antibodies were diluted in 0.5% non-fat dry milk in PBS and sections were stained at 4°C overnight in the dark. Sections were washed three times with PBS-X followed by three times with PBS. Secondary antibodies were diluted in 0.5% non-fat dry milk and sections were stained at room temperature for 2 h in the dark. Sections were washed three times with PBS-X followed by three times with PBS. X34 was diluted at 1:1,000 in staining buffer (60% PBS, 40% EtOH, 1:500 10 N NaOH) and sections were stained for 20 min at room temperature in the dark. Sections were washed with staining buffer without X34 three times for 2 min each, followed by PBS twice for 5 min each. Slides were coverslipped with Vectashield mounting medium without DAPI.
Primary antibodies: mouse anti-human MOAB-2-Alexa488 (1:500, NBP2-13075AF488; Novus), rabbit anti-mouse LAMP1 (1:500, ab208943; Abcam), rabbit anti-mouse P2RY12 (1:400, AS-55043A; Anaspec), rabbit anti-mouse IBA1 (1:200, 019-19741; Wako), goat anti-mouse LGALS3 (1:100, AF1197; R&D Systems), rabbit anti-mouse GFAP (1:250, 12389; Cell Signaling), and rat anti-mouse CD68 (1:200, MCA1957; Bio-Rad).
Fluorescence microscope image acquisition
Slides were imaged on a Keyence BZ-X800 microscope using the 10×, 20×, 40×, and 100× objectives. For stains X34, MOAB-2, and LAMP1, 10× stitches of the hippocampus and overlying cortex, imaged in a single plane, were generated. GFAP images were acquired at 10× in a single plane, z stack P2RY12, CD68, and LGALS3 images were acquired at 10× and 20×, and z stack IBA1 and LGALS3 images were acquired at 100×. Z stacks for each stain were acquired with a constant number of slices and pitch and stitched into a single full-focus image with the BZ-X800 image software by selecting the default “standard image” setting.
Plaque and neuritic dystrophy analysis
Plaques and neuritic dystrophy were quantified in ImageJ. Plaques and dystrophy were detected by thresholding X34 or LAMP1 signal intensity after segmenting the hippocampus and overlying cortex as separate regions of interest. Threshold values were calibrated using a representative batch of images. Total plaque area, total tissue area, number of plaques, and average plaque size were recorded.
Microglia and astrocyte Sholl analysis
Analysis of activated microglia and astrocytes was performed in Qupath with a customized Sholl analysis pipeline. Amyloid plaques were detected by thresholding X34 signal intensity. Threshold values were calibrated using a representative batch of images. After thresholding, the plaques were separated and saved into discrete annotation objects, and LGALS3, CD68, or GFAP intensity or coverage was measured within each object. Next, all annotation objects were expanded by 5 μm using the object dilation plugin, and intensity values were once again measured.
Statistical analysis
Statistics were performed using GraphPad Prism with significance set at P < 0.05. All comparisons were performed between IgG and aducanumab groups within each sex. Normality was checked with the Anderson-Darling, D’Agostino and Pearson, and Shapiro–Wilk tests (for groups with at least N = 8), and Shapiro–Wilk normality test (for groups with N < 8). The former two tests have minimum sample sizes of N = 8 per group; thus, in groups with N < 8, only the Shapiro–Wilk test was used. For groups with at least N = 8, if any of the three tests suggested a non-normal distribution (P < 0.05), a Mann–Whitney test was used for a more conservative analysis.
If data were normally distributed, the variance was checked with an F-test. If variances were not significantly different, an unpaired two-tailed Student’s t test was used. If variances were significantly different, a Welch’s t test was used.
For bulk RNAseq analysis, statistics were performed in edgeR (see bulk RNAseq analysis methods section and code for additional details). Significance was set at P < 0.1 using false discovery rate (FDR)–corrected adjusted P values. Additional details are provided in figures and legends.
Figures
Timeline diagrams in Fig. 1 A and Fig. 4 A and dissociation schematic in Fig. S2 A were created with http://BioRender.com.
Online supplemental material
Fig. S1 shows an extended analysis of plaque and neuritic dystrophy during the acute phase. Fig. S2 shows an extended analysis of microglial activation during the acute phase. Fig. S3 shows an extended analysis of plaque and neuritic dystrophy during the washout phase. Fig. S4 shows an extended analysis of sex differences in bulk RNAseq data. Fig. S5 shows an extended analysis of global and peri-plaque microgliosis and astrogliosis. Table S1 contains the data underlying Fig. 1. Table S2 contains the data underlying Fig. 2. Table S3 contains the data underlying Fig. 3. Table S4 contains the data underlying Fig. 4. Table S5 contains the data underlying Fig. 5. Table S6 contains the data underlying Fig. 6. Table S7 contains the data underlying Fig. S1. Table S8 contains the data underlying Fig S2. Table S9 contains the data underlying Fig. S3. Table S10 contains the data underlying Fig. S4. Table S11 contains the data underlying Fig. S5.
Supplementary Material
shows data underlying Fig. 1.
shows data underlying Fig. 2.
shows data underlying Fig. 3.
shows data underlying Fig. 4.
shows data underlying Fig. 5.
shows data underlying Fig. 6.
shows data underlying Fig. S1.
shows data underlying Fig. S2.
shows data underlying Fig. S3.
shows data underlying Fig. S4.
shows data underlying Fig. S5.
Acknowledgments
We would like to thank the Mayo Clinic Arizona Flow Cytometry Core, Mayo Clinic Rochester Genome Analysis Core, and Genome Analysis Core director Dr. Eric Weiben.
J.D. Fryer was supported by the following funding sources: Mayo Clinic Foundation, Ben Dov Family Luminescence Foundation, Ed and Ethel Moore Alzheimer’s Disease Research Program of Florida Department of Health (6AZ06), The Rotary Coins for Alzheimer’s Research Trust Fund, CureAlz Foundation, Goodman Family Foundation, and National Institutes of Health grants NS084974, AG062556, AG062110, NS094137, AG057997, AG062077, NS110435, AG047327, AG049992, and AG062677.
Author contributions: Methodology: M.P. Cadiz, J.D. Fryer, K.T. Todd, D.G. Nascari, K.C. Olney, M.M. Al-Amin. Investigation: M.P. Cadiz, K.A. Gibson, N. Massa, M.T. Lilley. Formal analysis: M.P. Cadiz, D.G. Nascari, K.T. Todd, K.C. Olney. Visualization: M.P. Cadiz, K.T. Todd. Funding acquisition: J.D. Fryer. Project administration: M.P. Cadiz, J.D. Fryer. Resources: J.D. Fryer, D.M. Holtzman, H. Jiang. Supervision: J.D. Fryer. Writing—original draft: M.P. Cadiz, J.D. Fryer. Writing—review and editing: M.P. Cadiz, K.A. Gibson, K.T. Todd, D.G. Nascari, N. Massa, M.T. Lilley, K.C. Olney, M.M. Al-Amin, H. Jiang, D.M. Holtzman, and J.D. Fryer.
Data availability
Data are available in the article itself and its supplementary materials. Bulk and scRNAseq data files are deposited in SRA (https://www.ncbi.nlm.nih.gov/sra?term=SRP465463). Code for bulk and scRNAseq are available at https://github.com/fryerlab/aducanumab.
References
- Andrews, S. Babraham Bioinformatics. 2010. FastQC: A quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Barrachina, M., Maes T., Buesa C., and Ferrer I.. 2006. Lysosome-associated membrane protein 1 (LAMP-1) in Alzheimer’s disease. Neuropathol. Appl. Neurobiol. 32:505–516. 10.1111/j.1365-2990.2006.00756.x [DOI] [PubMed] [Google Scholar]
- Benzing, W.C., Mufson E.J., and Armstrong D.M.. 1993. Alzheimer’s disease-like dystrophic neurites characteristically associated with senile plaques are not found within other neurodegenerative diseases unless amyloid β-protein deposition is present. Brain Res. 606:10–18. 10.1016/0006-8993(93)91563-8 [DOI] [PubMed] [Google Scholar]
- Braak, H., Braak E., Ohm T., and Bohl J.. 1989. Alzheimer’s disease: Mismatch between amyloid plaques and neuritic plaques. Neurosci. Lett. 103:24–28. 10.1016/0304-3940(89)90479-5 [DOI] [PubMed] [Google Scholar]
- Bushnell, B. 2014. BBMap: A Fast, Accurate, Splice-Aware Aligner. Lawrence Berkeley National Lab. LBNL, Berkeley, CA. [Google Scholar]
- Chen, Y., and Colonna M.. 2021. Microglia in Alzheimer’s disease at single-cell level. Are there common patterns in humans and mice? J. Exp. Med. 218:e20202717. 10.1084/jem.20202717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna, M., and Butovsky O.. 2017. Microglia function in the central nervous system during health and neurodegeneration. Annu. Rev. Immunol. 35:441–468. 10.1146/annurev-immunol-051116-052358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condello, C., Yuan P., Schain A., and Grutzendler J.. 2015. Microglia constitute a barrier that prevents neurotoxic protofibrillar Aβ42 hotspots around plaques. Nat. Commun. 6:6176. 10.1038/ncomms7176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings, B.J., Su J.H., Geddes J.W., Van Nostrand W.E., Wagner S.L., Cunningham D.D., and Cotman C.W.. 1992. Aggregation of the amyloid precursor protein within degenerating neurons and dystrophic neurites in Alzheimer’s disease. Neuroscience. 48:763–777. 10.1016/0306-4522(92)90265-4 [DOI] [PubMed] [Google Scholar]
- Da Mesquita, S., Papadopoulos Z., Dykstra T., Brase L., Farias F.G., Wall M., Jiang H., Kodira C.D., de Lima K.A., Herz J., et al. 2021. Meningeal lymphatics affect microglia responses and anti-Aβ immunotherapy. Nature. 593:255–260. 10.1038/s41586-021-03489-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson, D.W., Wertkin A., Mattiace L.A., Fier E., Kress Y., Davies P., and Yen S.-H.. 1990. Ubiquitin immunoelectron microscopy of dystrophic neurites in cerebellar senile plaques of Alzheimer’s disease. Acta Neuropathol. 79:486–493. 10.1007/BF00296107 [DOI] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellwanger, D.C., Wang S., Brioschi S., Shao Z., Green L., Case R., Yoo D., Weishuhn D., Rathanaswami P., Bradley J., et al. 2021. Prior activation state shapes the microglia response to antihuman TREM2 in a mouse model of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 118:e2017742118. 10.1073/pnas.2017742118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels, P., Magnusson M., Lundin S., and Käller M.. 2016. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 32:3047–3048. 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, B.A., Srinivasan K., Ayalon G., Meilandt W.J., Lin H., Huntley M.A., Cao Y., Lee S.-H., Haddick P.C.G., Ngu H., et al. 2018. Diverse brain myeloid expression profiles reveal distinct microglial activation states and aspects of Alzheimer’s disease not evident in mouse models. Cell Rep. 22:832–847. 10.1016/j.celrep.2017.12.066 [DOI] [PubMed] [Google Scholar]
- García-Revilla, J., Boza-Serrano A., Espinosa-Oliva A.M., Soto M.S., Deierborg T., Ruiz R., de Pablos R.M., Burguillos M.A., and Venero J.L.. 2022. Galectin-3, a rising star in modulating microglia activation under conditions of neurodegeneration. Cell Death Dis. 13:628. 10.1038/s41419-022-05058-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George, C., Gontier G., Lacube P., François J.-C., Holzenberger M., and Aïd S.. 2017. The Alzheimer’s disease transcriptome mimics the neuroprotective signature of IGF-1 receptor-deficient neurons. Brain. 140:2012–2027. 10.1093/brain/awx132 [DOI] [PubMed] [Google Scholar]
- Gowrishankar, S., Yuan P., Wu Y., Schrag M., Paradise S., Grutzendler J., De Camilli P., and Ferguson S.M.. 2015. Massive accumulation of luminal protease-deficient axonal lysosomes at Alzheimer’s disease amyloid plaques. Proc. Natl. Acad. Sci. USA. 112:E3699–E3708. 10.1073/pnas.1510329112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubman, A., Choo X.Y., Chew G., Ouyang J.F., Sun G., Croft N.P., Rossello F.J., Simmons R., Buckberry S., Landin D.V., et al. 2021. Transcriptional signature in microglia associated with Aβ plaque phagocytosis. Nat. Commun. 12:3015. 10.1038/s41467-021-23111-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, T.R., Dufort C., Dissing-Olesen L., Giera S., Young A., Wysoker A., Walker A.J., Gergits F., Segel M., Nemesh J., et al. 2019. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity. 50:253–271.e6. 10.1016/j.immuni.2018.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, Y., Hao S., Andersen-Nissen E., Mauck W.M. III, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. 2021. Integrated analysis of multimodal single-cell data. Cell. 184:3573–3587.e29. 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassiotis, S., Manavis J., Blumbergs P.C., Hattersley K.J., Carosi J.M., Kamei M., and Sargeant T.J.. 2018. Lysosomal LAMP1 immunoreactivity exists in both diffuse and neuritic amyloid plaques in the human hippocampus. Eur. J. Neurosci. 47:1043–1053. 10.1111/ejn.13913 [DOI] [PubMed] [Google Scholar]
- He, B., Perez S.E., Lee S.H., Ginsberg S.D., Malek-Ahmadi M., and Mufson E.J.. 2020. Expression profiling of precuneus layer III cathepsin D-immunopositive pyramidal neurons in mild cognitive impairment and Alzheimer’s disease: Evidence for neuronal signaling vulnerability. J. Comp. Neurol. 528:2748–2766. 10.1002/cne.24929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, S.S., Ebbert M.T.W., Baker K.E., Cook C., Wang X., Sens J.P., Kocher J.-P., Petrucelli L., and Fryer J.D.. 2018. Microglial translational profiling reveals a convergent APOE pathway from aging, amyloid, and tau. J. Exp. Med. 215:2235–2245. 10.1084/jem.20180653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul, H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T.K., David E., Baruch K., Lara-Astaiso D., Toth B., et al. 2017. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell. 169:1276–1290.e17. 10.1016/j.cell.2017.05.018 [DOI] [PubMed] [Google Scholar]
- Konishi, H., Okamoto T., Hara Y., Komine O., Tamada H., Maeda M., Osako F., Kobayashi M., Nishiyama A., Kataoka Y., et al. 2020. Astrocytic phagocytosis is a compensatory mechanism for microglial dysfunction. EMBO J. 39:e104464. 10.15252/embj.2020104464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasemann, S., Madore C., Cialic R., Baufeld C., Calcagno N., El Fatimy R., Beckers L., O’Loughlin E., Xu Y., Fanek Z., et al. 2017. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity. 47:566–581.e9. 10.1016/j.immuni.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law, C.W., Chen Y., Shi W., and Smyth G.K.. 2014. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15:R29. 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng, F., and Edison P.. 2021. Neuroinflammation and microglial activation in Alzheimer disease: Where do we go from here? Nat. Rev. Neurol. 17:157–172. 10.1038/s41582-020-00435-y [DOI] [PubMed] [Google Scholar]
- Li, Q., Cheng Z., Zhou L., Darmanis S., Neff N.F., Okamoto J., Gulati G., Bennett M.L., Sun L.O., Clarke L.E., et al. 2019. Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron. 101:207–223.e10. 10.1016/j.neuron.2018.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y., Smyth G.K., and Shi W.. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Mathys, H., Adaikkan C., Gao F., Young J.Z., Manet E., Hemberg M., De Jager P.L., Ransohoff R.M., Regev A., and Tsai L.-H.. 2017. Temporal tracking of microglia activation in neurodegeneration at single-cell resolution. Cell Rep. 21:366–380. 10.1016/j.celrep.2017.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon, R.A. 2007. Autophagy, amyloidogenesis and Alzheimer disease. J. Cell Sci. 120:4081–4091. 10.1242/jcs.019265 [DOI] [PubMed] [Google Scholar]
- Olney, K.C., Todd K.T., Pallegar P.N., Jensen T.D., Cadiz M.P., Gibson K.A., Barnett J.H., de Ávila C., Bouchal S.M., Rabichow B.E., et al. 2022. Widespread choroid plexus contamination in sampling and profiling of brain tissue. Mol. Psychiatry. 27:1839–1847. 10.1038/s41380-021-01416-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onorato, M., Mulvihill P., Connolly J., Galloway P., Whitehouse P., and Perry G.. 1989. Alteration of neuritic cytoarchitecture in Alzheimer disease. Prog. Clin. Biol. Res. 317:781–789. [PubMed] [Google Scholar]
- Rangaraju, S., Dammer E.B., Raza S.A., Rathakrishnan P., Xiao H., Gao T., Duong D.M., Pennington M.W., Lah J.J., Seyfried N.T., and Levey A.I.. 2018. Identification and therapeutic modulation of a pro-inflammatory subset of disease-associated-microglia in Alzheimer’s disease. Mol. Neurodegener. 13:24. 10.1186/s13024-018-0254-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., and Smyth G.K.. 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala Frigerio, C., Wolfs L., Fattorelli N., Thrupp N., Voytyuk I., Schmidt I., Mancuso R., Chen W.-T., Woodbury M.E., Srivastava G., et al. 2019. The major risk factors for Alzheimer’s disease: Age, sex, and genes modulate the microglia response to Aβ plaques. Cell Rep. 27:1293–1306.e6. 10.1016/j.celrep.2019.03.099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian Monasor, L., Müller S.A., Colombo A.V., Tanrioever G., König J., Roth S., Liesz A., Berghofer A., Piechotta A., Prestel M., et al. 2020. Fibrillar Aβ triggers microglial proteome alterations and dysfunction in Alzheimer mouse models. Elife. 9:e54083. 10.7554/eLife.54083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevigny, J., Chiao P., Bussière T., Weinreb P.H., Williams L., Maier M., Dunstan R., Salloway S., Chen T., Ling Y., et al. 2016. The antibody aducanumab reduces Aβ plaques in Alzheimer’s disease. Nature. 537:50–56. 10.1038/nature19323 [DOI] [PubMed] [Google Scholar]
- Song, W.M., and Colonna M.. 2018. The identity and function of microglia in neurodegeneration. Nat. Immunol. 19:1048–1058. 10.1038/s41590-018-0212-1 [DOI] [PubMed] [Google Scholar]
- Spangenberg, E., Severson P.L., Hohsfield L.A., Crapser J., Zhang J., Burton E.A., Zhang Y., Spevak W., Lin J., Phan N.Y., et al. 2019. Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer’s disease model. Nat. Commun. 10:3758. 10.1038/s41467-019-11674-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling, R.A., Jack C.R. Jr., Black S.E., Frosch M.P., Greenberg S.M., Hyman B.T., Scheltens P., Carrillo M.C., Thies W., Bednar M.M., et al. 2011. Amyloid-related imaging abnormalities in amyloid-modifying therapeutic trials: Recommendations from the Alzheimer’s association research roundtable workgroup. Alzheimers Dement. 7:367–385. 10.1016/j.jalz.2011.05.2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., Mustafa M., Yuede C.M., Salazar S.V., Kong P., Long H., Ward M., Siddiqui O., Paul R., Gilfillan S., et al. 2020. Anti-human TREM2 induces microglia proliferation and reduces pathology in an Alzheimer’s disease model. J. Exp. Med. 217:e20200785. 10.1084/jem.20200785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcock, D.M., Gordon M.N., Ugen K.E., Gottschall P.E., DiCarlo G., Dickey C., Boyett K.W., Jantzen P.T., Connor K.E., Melachrino J., et al. 2001. Number of Abeta inoculations in APP+PS1 transgenic mice influences antibody titers, microglial activation, and congophilic plaque levels. DNA Cell Biol. 20:731–736. 10.1089/10445490152717596 [DOI] [PubMed] [Google Scholar]
- Wilcock, D.M., DiCarlo G., Henderson D., Jackson J., Clarke K., Ugen K.E., Gordon M.N., and Morgan D.. 2003. Intracranially administered anti-Abeta antibodies reduce β-amyloid deposition by mechanisms both independent of and associated with microglial activation. J. Neurosci. 23:3745–3751. 10.1523/JNEUROSCI.23-09-03745.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcock, D.M., Rojiani A., Rosenthal A., Levkowitz G., Subbarao S., Alamed J., Wilson D., Wilson N., Freeman M.J., Gordon M.N., and Morgan D.. 2004. Passive amyloid immunotherapy clears amyloid and transiently activates microglia in a transgenic mouse model of amyloid deposition. J. Neurosci. 24:6144–6151. 10.1523/JNEUROSCI.1090-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, M., Jiang H., Serrano J.R., Gonzales E.R., Wang C., Gratuze M., Hoyle R., Bien-Ly N., Silverman A.P., Sullivan P.M., et al. 2021. APOE immunotherapy reduces cerebral amyloid angiopathy and amyloid plaques while improving cerebrovascular function. Sci. Transl. Med. 13:eabd7522. 10.1126/scitranslmed.abd7522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., and Chanda S.K.. 2019. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10:1523. 10.1038/s41467-019-09234-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Song W.M., Andhey P.S., Swain A., Levy T., Miller K.R., Poliani P.L., Cominelli M., Grover S., Gilfillan S., et al. 2020. Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 26:131–142. 10.1038/s41591-019-0695-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows data underlying Fig. 1.
shows data underlying Fig. 2.
shows data underlying Fig. 3.
shows data underlying Fig. 4.
shows data underlying Fig. 5.
shows data underlying Fig. 6.
shows data underlying Fig. S1.
shows data underlying Fig. S2.
shows data underlying Fig. S3.
shows data underlying Fig. S4.
shows data underlying Fig. S5.
Data Availability Statement
Data are available in the article itself and its supplementary materials. Bulk and scRNAseq data files are deposited in SRA (https://www.ncbi.nlm.nih.gov/sra?term=SRP465463). Code for bulk and scRNAseq are available at https://github.com/fryerlab/aducanumab.