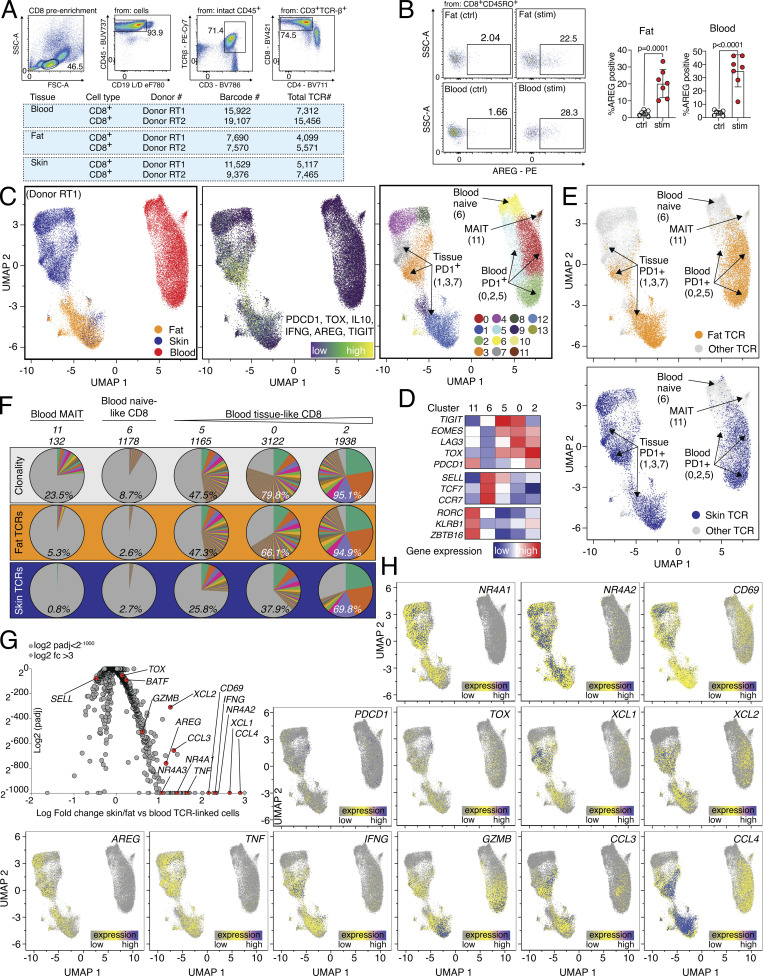
Figure S2.
scRNA/TCR-seq of human CD8 T cells with donor RT1. (A) Top, sort layout for CD8 pre-enriched immune cells from human blood. Bottom, QC of cells used for combined scRNA/TCR-seq isolated from human blood, fat, and skin. (B) Gating and quantification of AREG expression in human fat (left) and blood (right). CD8 T cells after o/n stimulation with TI (Ctrl) or PMA/ionomycin in the presence of TI (stim), n = 7. (C and D) Left, cells color-coded based on tissue of origin. Middle, expression of gene signature (PDCD1, TOX, IL10, IFNG, AREG, and TIGIT). Right, cells clustered in 12 groups. Annotation of clusters using signature genes shown in (D) and labeled in UMAP. (E) Top, TCRs derived from all fat CD8 T cells in cluster 1 (6,381 cells) are highlighted in yellow and displayed in all other clusters. Bottom, TCRs derived from all skin CD8 T cells in clusters 3,7 (8,627 cells) highlighted in blue and displayed in all other clusters. (F) Clonality of clusters (top, white), or tracking of fat (middle, orange) or skin (bottom, blue) CD8 T cells in blood-based clusters of the same donor. The percentage indicates the fraction of detected clones among total clones for the donor, with the total number of clones shown above. Each slice represents a clonotype with the angle representing its fraction among all cells in the respective cluster. (G) DEGs between TCR-identical cells in clusters 1, 3, 7 and 0, 2, 5, 6, 11. Several genes highlighted in red and labeled, Padj values <2−1000 capped at 2−1000. Values >3 capped at 3. (H) Gene expression of NR4A1, NR4A2, CD69, TOX, XCL1, XCL2, AREG, TNF, IFNG, GZMB, CCL3, PDCD1, and CCL4 in CD8 T cells from donor RT1. CDR3 sequences are listed in Table S4. All data are derived from two or more independent experiments with an additional donor RT2 shown in Fig. 3.