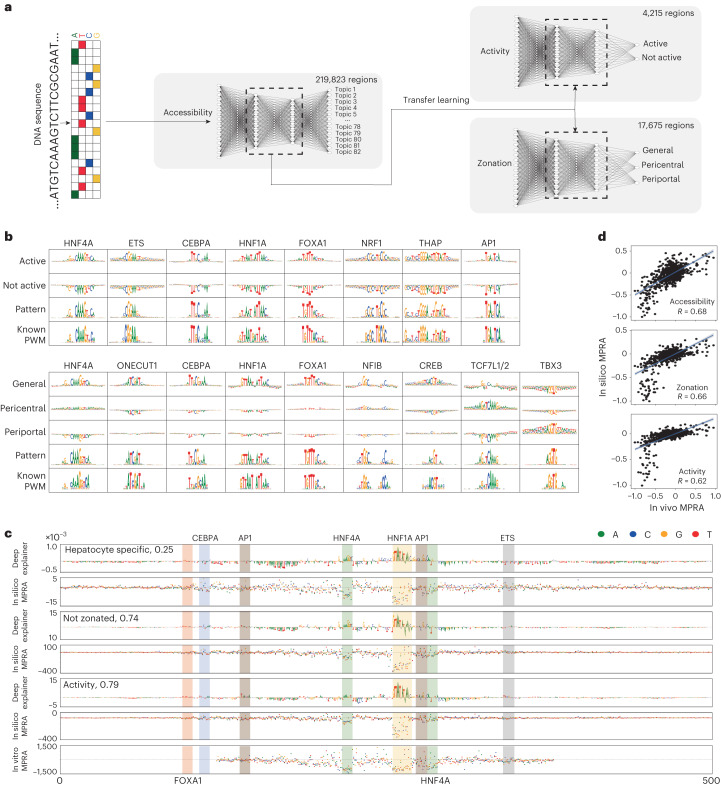
Fig. 4. DeepLiver decodes enhancer grammar.
a, DeepLiver overview. First, a CNN is trained to classify DNA sequences into their corresponding regulatory topic (219,823 sequences). The weights learned in the first model are used to initialize the activity and zonation models. The activity model classifies DNA sequences on the basis of their MPRA activity in vivo (using 4,215 high-confidence regions), while the zonation model classifies sequences on the basis of their zonation pattern on hepatocytes (pericentral, periportal or non zonated/general, using 17,675 regions for training). b, TF-MoDISco patterns identified in the activity and zonation models, with their contribution score per class and their most similar PWM from the cisTarget motif collection. c, DeepExplainer and saturation mutagenesis plots for the accessibility, zonation and activity models on an Aldob enhancer (hg19: chromosome 9: 104195449–104195449), with motifs highlighted. Saturation mutagenesis, shown below, was performed in this enhancer previously41. d, The correlation between DeepLiver in silico mutagenesis and experimental saturation mutagenesis in the Aldob enhancer. The blue line represents the fitted linear regression and the grey bands represent the 95% confidence interval bands. Source numerical data are provided as source data.