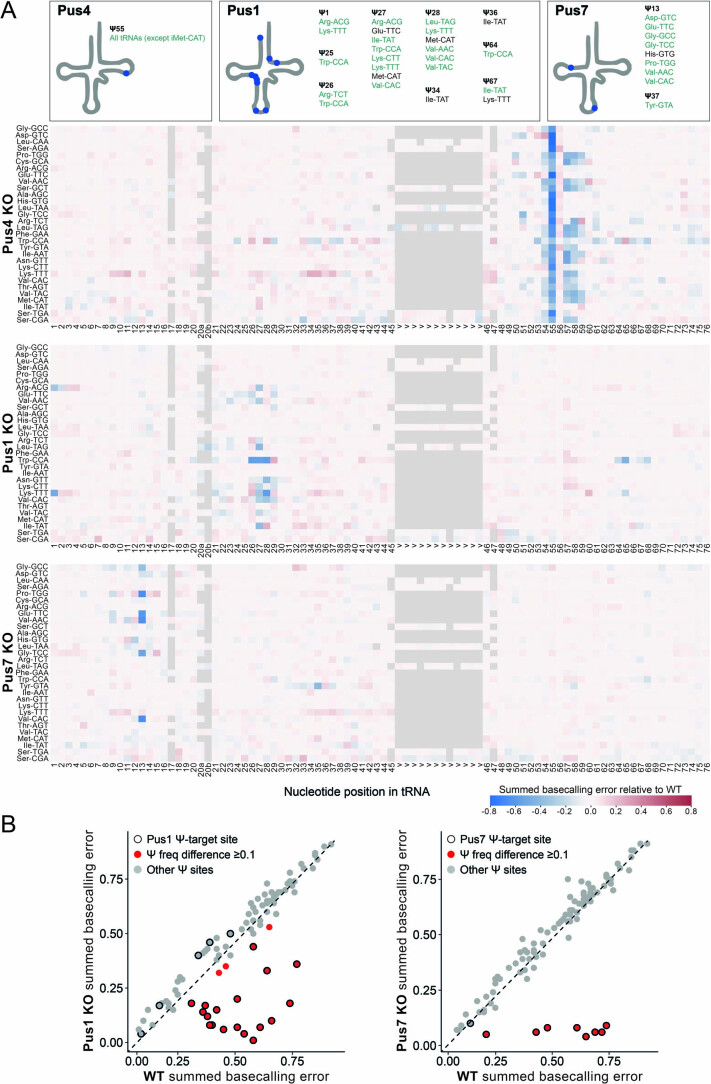
Extended Data Fig. 9. Nano-tRNAseq can capture RNA modifications changes upon knockout of pseudouridine synthase enzymes.
(a) Heatmap of summed basecalling error frequency of Pus4 KO biological replicate 2, Pus1 KO, and Pus7 KO (see also Supplementary Table 16 and Supplementary Table 17). The known positions of specific RNA modifications in each tRNA, as listed in MODOMICS, are shown in schematic above, as well as listed in Supplementary Table 18. Ψ positions observed in Nano-tRNAseq are highlighted in green (greater or equal to 0.1, see Supplementary Table 15). Nucleotides with a higher summed basecalling error frequency relative to WT are in red tones, and those with a lower summed basecalling error frequency are in blue tones. (b) Comparison of mismatch frequencies for known Ψ sites in S. cerevisiae WT vs. Pus1 and Pus7 KO tRNA molecules. Each data point represents a known tRNA Ψ site; a black outline indicates Ψ55 sites targeted by the enzyme in question, and a red fill indicates sites with a summed basecalling error of ≥0.1 for Pus 1 KO and Pus 7 KO compared to WT, which serves as a proxy for Ψ modification frequency.