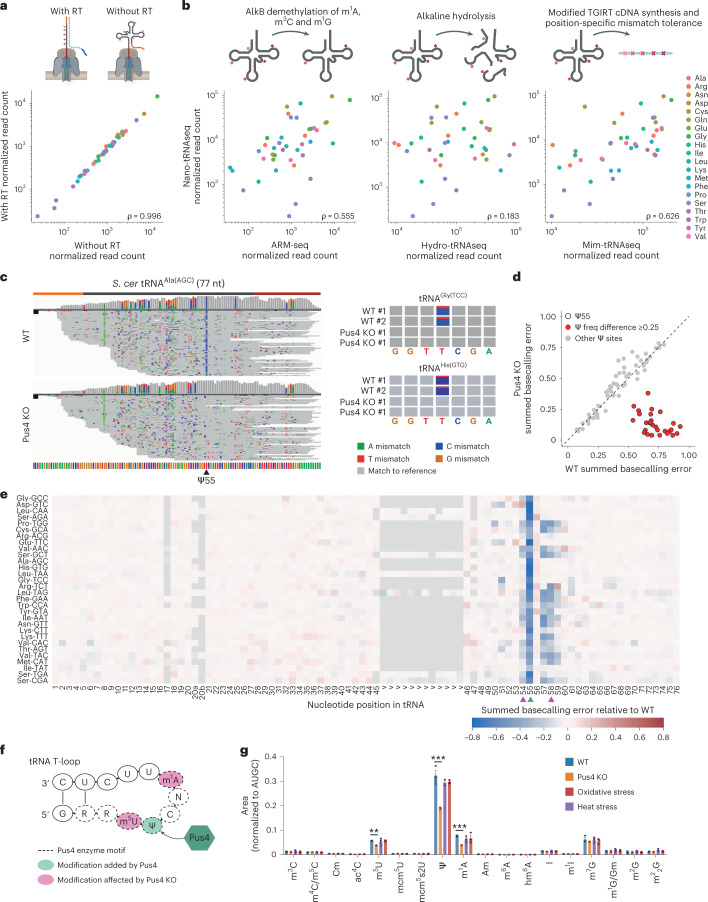
Fig. 4. Nano-tRNAseq can quantify tRNA abundance and RNA modifications as well as capture modification interdependencies.
a,b, Scatter plots of WT S. cerevisiae tRNA abundances sequenced with Nano-tRNAseq with and without the reverse transcription step (a) and compared to orthogonal Illumina-based tRNA sequencing methods (b). Each point represents a tRNA alloacceptor and is colored by alloacceptor type; the key is shown in b. The correlation strength is indicated by Spearman’s correlation coefficient (ρ). c, IGV tracks of tRNAAla(AGC) from WT and Pus4 KO S. cerevisiae strains (n = 2 biological replicates). Ψ55 is indicated with a black arrowhead. Adjacent are zoomed IGV snapshots of the Ψ55 region. Positions with a mismatch frequency greater than 0.2 are colored, whereas those lower than 0.2 are shown in gray. d, Scatter plot showing the mismatch frequencies for Ψ sites in S. cerevisiae WT versus Pus4 KO tRNA molecules. Each data point represents a known tRNA Ψ site; a black outline indicates Ψ55 sites; and a red fill indicates sites with a summed basecalling error of ≥0.25 compared to WT. e, Heat map of the summed basecalling error of Pus4 KO relative to WT, for each nucleotide (x axis) and for each tRNA isoacceptor (y axis, ordered from most to least abundant in descending order)(Supplementary Table 17). The positions of known tRNA modifications found in each tRNA gene are listed in Supplementary Table 18. The Pus4 target Ψ55 is indicated with a green arrowhead and m5U54 and m1A58 with pink arrowheads. See also Extended Data Fig. 9a for biological replicate 2. f, Schematic of the tRNA T-loop targeted by the Pus4 enzyme. Nucleotides with a dotted outline represent the Pus4 binding motif (RRUUCNA); Ψ55 is highlighted in green; and m5U54 and m1A58 are highlighted in pink. g, LC–MS/MS validation of S. cerevisiae tRNA modification levels. See also Supplementary Tables 19 and 20. Bars represent mean ± s.e.m. for n = 3 biological replicates per condition. P values were determined using a one-way ANOVA with Tukey correction for multiple comparisons, and significance was assessed by comparison to WT. *P < 0.05, **P < 0.01, ***P < 0.001, P(m5U) = 0.0015, P(Ψ) and P(m1A) < 0.0001. RT, reverse transcription.