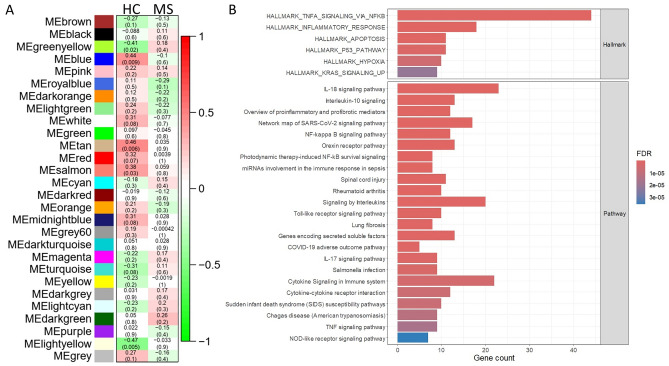
Figure 6.
Vitamin D-regulated pathways in CD4+ T cells determined through weighted gene co-expression network analysis. (A) Consensus modules of healthy control (HC) and multiple sclerosis (MS) groups. For each module, the number represents the correlation coefficient between their respective module eigengene (ME) and plasma 25(OH)D level, and the number in brackets represents the P-value. (B) Functional enrichment results for the “light yellow” module, with the top 30 enriched Hallmark gene sets and pathways shown. X-axis represents the number of genes of interest present in the annotated term, and bar color represents the false discovery rate (FDR).