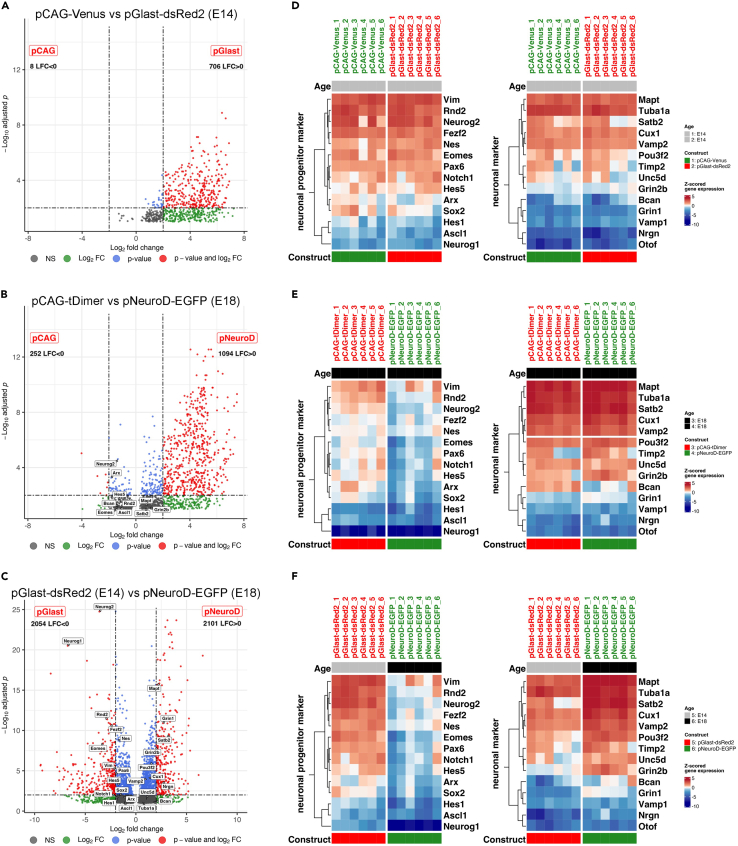
Figure 7.
Transcriptional analysis in progenitors and developing neurons reveal differentially expressed genes (DEGs) and enrichment for developmentally regulated marker gene expression in specific progenitor and neuronal subpopulations
(A–C) Flow cytometry-based isolation of cell populations sorted by fluorescent proteins driven by developmentally active promoters show enrichment for characteristic genes of the respective targeted cell identity (white boxes) after bulk RNA-seq. A) At E14, DEGs were detected in pGlast-dsRed2 positive progenitors compared to pCAG-Venus positive controls, but no enrichment for characteristic genes of progenitor cell identity was observed. B) DEGs were detected in isolated pNeuroD-eGFP positive immature neurons compared to pCAG-tDimer positive controls at E18. A volcano plot shows enrichment of multiple individual genes involved in neuronal developmental indicating enrichment of characteristic genes of early neuronal cell identity (white boxes). c) DEGs were detected in pGlast-dsRed2 positive progenitors (E14) compared to pNeuroD-eGFP positive developing neurons (E18). A volcano plot shows enrichment of multiple individual genes indicating enrichment of characteristic genes of progenitor cell identity in pGlast-dsRed2 positive cells (E14) and enrichment of several individual genes involved in neuronal development in pNeuroD-eGFP positive immature neurons (E18), confirming progenitor and neuronal cell identity, respectively (white boxes). Gray = expression not significantly altered, green = Log2 fold change (FC), blue = p-value, red = Log2 fold change and p-value: Genes plotted in red passed the p.adj. cut-off of <0.01 and the Log2 FC cut-off >2.00.
(D–F) Commonly used marker genes for progenitor (left) and developing neuronal cells (right), respectively, identified in bulk RNA-seq validate differences in cell identity of isolated cells labeled with D) pCAG-Venus vs. pGlast-dsRed2, E) pNeuroD-eGFP vs. pCAG-tDimer or f) pGlast-dsRed2 vs. pNeuroD-eGFP at time points E14 (gray bars) and E18 (black bars). References to the common usage of these genes in RNA-seq analyses are stated in the expected outcomes section. To facilitate comparison, pGlast (E14) and pNeuroD (E18) samples (in F) compare the same input from the cotransfected cell populations, which were independently scored against one another. Z-score was applied on normalized counts (using the DESeq2::rlog method); heatmap colors range from red (high expression), to white (moderate expression) to blue (low expression).