Abstract
The genus Fusarium includes numerous important plant and human pathogens, as well as many industrially and commercially important species. During our investigation of fungal diversity in China, a total of 356 fusarioid isolates were obtained and identified from diverse diseased and healthy plants, or different environmental habitats, i.e., air, carbonatite, compost, faeces, soil and water, representing hitherto one of the most intensive sampling and identification efforts of fusarioid taxa in China. Combining morphology, multi-locus phylogeny and ecological preference, these isolates were identified as 72 species of Fusarium and allied genera, i.e., Bisifusarium (1), Fusarium (60), and Neocosmospora (11). A seven-locus dataset, comprising the 5.8S nuclear ribosomal RNA gene with the two flanking internal transcribed spacer (ITS) regions, the intergenic spacer region of the rDNA (IGS), partial translation elongation factor 1-alpha (tef1), partial calmodulin (cam), partial RNA polymerase largest subunit (rpb1), partial RNA polymerase second largest subunit (rpb2) gene regions, and partial β-tubulin (tub2), were sequenced and employed in phylogenetic analyses. A genus-level phylogenetic tree was constructed using combined tef1, rpb1, and rpb2 sequences, which confirmed the presence of four fusarioid genera among the isolates studied. Further phylogenetic analyses of two allied genera (Bisifusarium and Neocosmospora) and nine species complexes of Fusarium were separately conducted employing different multi-locus datasets, to determine relationships among closely related species. Twelve novel species were identified and described in this paper. The F. babinda species complex is herein renamed as the F. falsibabinda species complex, including descriptions of new species. Sixteen species were reported as new records from China.
Citation: Wang MM, Crous PW, Sandoval-Denis M, et al. 2022. Fusarium and allied genera from China: species diversity and distribution.
Persoonia 48: 1-53. https://doi.org/10.3767/persoonia.2022.48.01.
Keywords: Fusarium, multigene phylogeny, new taxa, species complex, systematics
INTRODUCTION
Fusarium and allied genera comprise a large number of destructive pathogens and mycotoxigenic fungi, threatening plant, animal, and human health, as well as food security (O’Donnell et al. 2013). Fusarioid species cause many notable plant diseases, such as Fusarium head blight or scab of cereals by members of the F. graminearum species complex (O’Donnell et al. 2000a, Cuomo et al. 2007), sudden death syndrome of soybeans (Aoki et al. 2005) and root rot of many diverse hosts (Coleman et al. 2009, Sandoval-Denis et al. 2018a) by members of Neocosmospora (Lombard et al. 2015, Sandoval-Denis et al. 2019), ear rot of maize by members of the F. fujikuroi species complex (Desjardins et al. 2002), and vascular wilts of many economically important crops by members of the F. oxysporum species complex (O’Donnell et al. 1998b, Skovgaard et al. 2001, Van der Does et al. 2008, Lombard et al. 2019a, Maryani et al. 2019a). Species within this complex are also well-known for their ability to produce a range of secondary metabolites, including some notorious mycotoxins produced by Fusarium spp. in cereals (Marasas et al. 1984, O’Donnell et al. 2018).
Since the establishment of Fusarium (Link 1809), the taxonomic framework of this genus has undergone several significant changes. Link (1809) determined the primary morphological character of Fusarium to be the distinctive canoe- or banana-shaped conidia. Wollenweber & Reinking (1935) surveyed the morphology of macro- and microconidia, and the presence of chlamydospores, sclerotia and sporodochia, and suggested that Fusarium should be divided into 16 morphological sections, including 65 species and 77 varieties and forms. In the next several decades, this system has largely influenced subsequent taxonomic studies. Despite the impact of this system, other several controversial viewpoints persisted. Snyder & Hansen (1940, 1941, 1945, 1954) reduced the number of species to nine with a number of formae speciales, and highlighted the importance of morphological observations based on cultures derived from single-spore isolates. Gordon (1944, 1952, 1954a, b, 1956a, b, 1959, 1960) developed a pragmatic approach that combined sexual morph morphology, incorporating some thoughts from Wollenweber & Reinking (1935) and Snyder & Hansen (1940, 1941, 1945, 1954), which accepted 26 species in the genus. Booth (1971) introduced the morphology of the conidiogenous cells as a species-level diagnostic character. Nelson et al. (1983) provided a detailed morphological identification manual for Fusarium. Nevertheless, the species identification in Fusarium, based on morphology was still confusing, because of the variable phenotypes in culture, intricate or too vague descriptions of species among different studies, and the historically complicated subspecies level ranks (Leslie & Summerell 2006, Lombard et al. 2019a, b, Wang et al. 2019).
During the last three decades, phylogenetic inference played an increasingly important role in Fusarium taxonomy (Lombard et al. 2019a, b). Many morphological sections in the system of Wollenweber & Reinking (1935), e.g., sections Discolor and Elegans, proved to be polyphyletic based on rpb1-rpb2 analyses (O’Donnell et al. 2013). Debates about the generic boundary of Fusarium also led to disagreement among taxonomists. Gräfenhan et al. (2011) and Schroers et al. (2011) introduced several genera in the basal Fusarium clade in the Nectriaceae, and indicated that several monophyletic clusters in the terminal Fusarium clade corresponded to other genera, including Neocosmospora. However, Geiser et al. (2013) insisted on a broader definition of Fusarium, to avoid the introduction of additional genera. By means of a 10-locus phylogenetic analysis, Lombard et al. (2015) delineated several genera in the terminal Fusarium clade, e.g., Bisifusarium (F. dimerum species complex), Rectifusarium (F. ventricosum species complex), and also resurrected some older generic names, e.g., Albonectria (F. decemcellulare species complex), and Neocosmospora (F. solani species complex). Based on the combined ITS-LSU-rpb1-rpb2-tef1 dataset, Crous et al. (2021) re-examined the fusarioid taxa in Nectriaceae and showed that the Wollenweber concept of Fusarium presently encompasses 20 distinct genera, including four new genera (Luteonectria, Nothofusarium, Scolecofusarium, and Setofusarium). Following the end of dual nomenclature, the genus Fusarium as currently circumscribed accommodates members that belong to the Gibberella clade (O’Donnell et al. 2013, Lombard et al. 2015), including 18 species complexes (Laurence et al. 2011, Aoki et al. 2014, O’Donnell et al. 2013, Zhou et al. 2016, Sandoval-Denis et al. 2018a, Lombard et al. 2019a, Crous et al. 2021). Numerous cryptic species have recently been uncovered based on multi-locus phylogeny, morphology, and ecological characteristics (Gordon & Martyn 1997, O’Donnell et al. 2000a, b, 2008, 2009a, b, Laurence et al. 2014, Lombard et al. 2019a, b, Sandoval-Denis et al. 2018a, b, 2019, Maryani et al. 2019a, b, Wang et al. 2019, Xia et al. 2019, Yilmaz et al. 2021).
Previous investigations on Fusarium in China were summarised by Yu (1955), in which 77 species, varieties and formae speciales of pathogenic Fusarium spp. from 55 plant hosts were listed. A wider sampling region included 103 species, varieties and formae speciales of Fusarium and Gibberella (sexual morph of Fusarium s.str.) obtained from at least 111 plant species, faeces, and soil (Tai 1979). Identifications in both studies employed the morphology-based taxonomic system of Wollenweber & Reinking (1935). According to the currently used taxonomic system, only 31 of the 77 names in Yu (1955), and 36 of the 103 names in Tai (1979) remain in Fusarium. Considering the importance of Fusarium and allied species, it is necessary to clarify the species diversity and distribution of Fusarium in China in a modern taxonomic framework.
In our continuous survey of phytopathogenic fungi from China, 356 fusarioid strains have been isolated from diverse plant materials and various environmental samples including air, carbonatite, compost, faeces, water, and soil. In this study through a combination of morphology, multi-locus phylogeny and ecological characteristics, we advanced our knowledge on the species diversity of fusarioid taxa from China, as well as their host range and distribution.
MATERIALS AND METHODS
Sample collection
Samples were collected from 15 provinces (Fujian, Guangdong, Guizhou, Hainan, Hebei, Hubei, Hunan, Jiangsu, Jiangxi, Qinghai, Shandong, Shanxi, Sichuan, Yunnan, and Zhejiang), three autonomous regions (Guangxi Zhuang, Neimenggu, and Tibet) and two municipalities (Beijing and Chongqing) in China, and isolated from agricultural products imported into China from 13 other countries (Argentina, Australia, Brazil, Canada, Italy, Japan, Netherlands, Philippines, Poland, Saudi Arabia, Spain, Ukraine, and USA). Diseased and healthy plant tissues were collected and placed in paper bags. Air samples were collected using the Koch sedimentation method (Zhang et al. 2017). Water samples were collected as 10 mL samples and kept in sterile 15 mL centrifuge tubes (Zhang et al. 2017). Compost, faeces, pollen, and soil samples were collected (10–100 g per sample) after removing the surface layer (Zhang et al. 2017). Carbonatite samples were collected as five pieces in different orientations at each sample site (Zhang et al. 2017).
Fungal isolation
Fungi were isolated from plant tissues using single spore isolation as outlined in Zhang et al. (2013). Fungal endophytes were isolated using a tissue isolation method. Briefly, plant tissue pieces (4–5 mm2) were taken from the margin of leaf or stem spots as well as healthy sections, consecutively immersed in 70 % ethanol for 1 min, 5 % NaClO for 3 min, 70 % ethanol for 1 min, and rinsed in sterile distilled water for 30 s. Tissue pieces were blotted dry in sterile paper towels and incubated on 1/4 strength potato dextrose agar (PDA; Crous et al. 2019) containing ampicillin and streptomycin (50 mg/L each) (Liu et al. 2015). Isolates were retrieved from compost, pollen, soil, and water using the plate dilution method. One gram of compost, faeces, pollen, soil, or water was suspended in 9 mL sterile water. The suspension was shaken on the Vortex vibration meter for 10 min. The extract was diluted to a series of concentrations, i.e., 10-2 to 10-5. For each concentration, 200 μL suspensions were spread onto the 1/4 strength potato dextrose agar (PDA) with three replicates. Carbonatite samples were treated following the protocol of Zhang et al. (2017).
All plates were incubated at room temperature and examined every 2 d for fungal hyphae. Individual colonies were picked up with a sterilised needle and transferred onto fresh PDA plates. All the cultures were then purified using an optimized protocol of single spore isolation (Zhang et al. 2013).
All isolates examined in this study were deposited in Lei Cai’s personal culture collection (LC), housed at the Institute of Microbiology, Chinese Academy of Sciences, Beijing, China. Information of isolates including geographic distribution and host/habitat are listed in Table 1. Type specimens of new species were deposited in the Mycological Fungarium of the Institute of Microbiology, Chinese Academy of Sciences, Beijing, China (HMAS), and living ex-type cultures in the China General Microbiological Culture Collection Centre (CGMCC).
Table 1.
Details of examined isolates included in the phylogenetic analyses. Newly generated GenBank accessions are in bold
| Species | Isolate | Country/Location | Host/Habitat | ITS | cam | tef1 | rpb1 | rpb2 | tub2 | IGS |
|---|---|---|---|---|---|---|---|---|---|---|
| Albonectria | ||||||||||
| A. rigidiuscula Bisifusarium | LC13606 = F503 | Japan | unidentified plant | MW016388 | MW566255 | MW580428 | MW024420 | MW474374 | MW533715 | – |
| B. aseptatum | CGMCC 3.20816 = LC1075 T | China, Guangdong Province, Guangzhou city | Orchidaceae sp. | MW016389 | MW566256 | MW580429 | MW024421 | MW474375 | MW533716 | – |
| LC13607 | China, Guangdong Province, Guangzhou city | Orchidaceae sp. | MW016390 | MW566257 | MW580430 | MW024422 | MW474376 | MW533717 | – | |
| LC13608 | China, Guangdong Province, Guangzhou city | Orchidaceae sp. | MW016391 | MW566258 | MW580431 | MW024423 | MW474377 | MW533718 | – | |
| Fusarium | ||||||||||
| F. concolor species complex | ||||||||||
| F. anguioides | LC13612 = M0563 | China, Guangdong Province, Shenzhen city | Cordyline stricta | MW016395 | MW566262 | MW580435 | MW024426 | MW474381 | MW533721 | – |
| LC13613 = M0568 | China, Guangdong Province, Shenzhen city | Alocasia odora | MW016396 | MW566263 | MW580436 | MW024427 | MW474382 | MW533722 | – | |
| LC7007 | China, Jiangxi Province | bamboo | MW016397 | MW566264 | MW580437 | MW024428 | MW474383 | – | – | |
| LC7151 | China, Jiangxi Province | bamboo | MW016398 | MW566265 | MW580438 | MW024429 | MW474384 | MW533723 | – | |
| LC7178 | China, Jiangxi Province | bamboo | MW016399 | MW566266 | MW580439 | MW024430 | MW474385 | MW533724 | – | |
| LC7189 | China, Guangdong Province, Guangzhou city | bamboo | MW016400 | MW566267 | MW580440 | MW024431 | MW474386 | MW533725 | – | |
| LC7190 | China, Guangdong Province, Guangzhou city | bamboo | MW016401 | MW566268 | MW580441 | MW024432 | MW474387 | – | – | |
| LC7240 | China, Jiangxi Province, Ganzhou city | bamboo | MW016402 | MW566269 | MW580442 | MW024433 | MW474388 | MW533726 | – | |
| F. bambusarum | CGMCC 3.20820 = LC7180 T | China, Jiangxi Province | bamboo | MW016403 | MW566270 | MW580443 | MW024434 | MW474389 | – | – |
| LC7187 | China, Guangdong Province, Guangzhou city | bamboo | MW016404 | MW566271 | MW580444 | MW024435 | MW474390 | – | – | |
| F. falsibabinda species complex | ||||||||||
| F. falsibabinda | CGMCC 3.20823 = LC13610 = F015 T | Japan | Podocarpus macrophyllus | MW016393 | MW566260 | MW580433 | MW024424 | MW474379 | MW533719 | – |
| LC13611 = F058 | Japan | Camellia sasanqua | MW016394 | MW566261 | MW580434 | MW024425 | MW474380 | MW533720 | – | |
| F. fujikuroi species complex | ||||||||||
| F. annulatum | LC1105 | China | Lithocarpus glabra | MW016472 | MW566339 | MW580512 | MW024500 | MW474458 | MW533791 | – |
| LC11490 = G2 | China, Beijing | Vitis sp. | MW016473 | MW566340 | MW580513 | MW024501 | MW474459 | MW533792 | – | |
| LC11527 = G358 | China, Hebei Province | Vitis sp. | MW016474 | MW566341 | MW580514 | MW024502 | MW474460 | MW533793 | – | |
| LC11584 = G373 | China, Hebei Province | Vitis sp. | MW016475 | MW566342 | MW580515 | MW024503 | MW474461 | MW533794 | – | |
| LC11650 = HM259-L09 | China, Hainan Province | Oryza sp. | MW016476 | MW566343 | MW580516 | MW024504 | MW474462 | MW533795 | – | |
| LC11670 = HM259-S07 | China, Hainan Province | Oryza sp. | MW016477 | MW566344 | MW580517 | MW024505 | MW474463 | MW533796 | – | |
| LC11672 = HM259-S12 | China, Hainan Province | Oryza sp. | MW016478 | MW566345 | MW580518 | MW024506 | MW474464 | MW533797 | – | |
| LC13658 = CF4 | China, Neimenggu Province | unidentified mushroom | MW016479 | MW566346 | MW580519 | MW024507 | MW474465 | MW533798 | – | |
| LC13659 = F007 | USA | Glycine max | MW016480 | MW566347 | MW580520 | MW024508 | MW474466 | MW533799 | – | |
| LC13660 = F023 | Philippines | Musa sp. | MW016481 | MW566348 | MW580521 | MW024509 | MW474467 | MW533800 | – | |
| LC13661 = F028 | Italy | Malus domestica | MW016482 | MW566349 | MW580522 | MW024510 | MW474468 | MW533801 | – | |
| LC13662 = F059 | Spain | Chamaerops humilis | MW016483 | MW566350 | MW580523 | MW024511 | MW474469 | MW533802 | – | |
| LC13663 = F100 | Ukraine | Zea mays | MW016484 | MW566351 | MW580524 | MW024512 | MW474470 | MW533803 | – | |
| LC13664 = F102 | USA | Sorghum bicolor | MW016485 | MW566352 | MW580525 | MW024513 | MW474471 | MW533804 | – | |
| LC13665 = F405 | Spain | Olea europaea | MW016486 | MW566353 | MW580526 | MW024514 | MW474472 | MW533805 | – | |
| LC13666 = GDBYL08-E1 | China, Guangdong Province, Guangzhou city | Musa nana | MW016487 | MW566354 | MW580527 | MW024515 | MW474473 | MW533806 | – | |
| LC13667 = GDBYL10-E1 | China, Guangdong Province, Guangzhou city | Musa nana | MW016488 | MW566355 | MW580528 | MW024516 | MW474474 | MW533807 | – | |
| LC13668 = GDGZSJL01E1 | China, Guangdong Province, Guangzhou city | Musa nana | MW016489 | MW566356 | MW580529 | MW024517 | MW474475 | MW533808 | – | |
| LC13669 = GXBSMAS2-E3 | China, Guangxi Zhuang Autonomous Region, Baise city | Musa nana | MW016490 | MW566357 | MW580530 | MW024518 | MW474476 | MW533809 | – | |
| LC13670 = GXCZMQS1-E2 | China, Guangxi Zhuang Autonomous Region, Chongzuo city | Musa nana | MW016491 | MW566358 | MW580531 | MW024519 | MW474477 | MW533810 | – | |
| LC13671 = GXLBL15-3 | China, Guangxi Zhuang Autonomous Region, Laibin city | Musa nana | MW016492 | MW566359 | MW580532 | MW024520 | MW474478 | MW533811 | – | |
| LC13673 = HBF3-2 | China, Hebei Province | Oryza sp. | MW016494 | MW566361 | MW580534 | MW024522 | MW474480 | MW533813 | – | |
| LC13674 = JXF3-22 | China, Jiangxi Province | Oryza sp. | MW016495 | MW566362 | MW580535 | MW024523 | MW474481 | MW533814 | – | |
| LC13675 = JXN1-21 | China, Jiangxi Province | Oryza sp. | MW016496 | MW566363 | MW580536 | MW024524 | MW474482 | MW533815 | – | |
| LC2825 | China, Beijing | unidentified grass | MW016497 | MW566364 | MW580537 | MW024525 | MW474483 | MW533816 | – | |
| LC5984 | China | submerged wood | MW016498 | MW566365 | MW580538 | MW024526 | MW474484 | MW533817 | – | |
| LC6002 | China | submerged wood | MW016499 | MW566366 | MW580539 | MW024527 | MW474485 | MW533818 | – | |
| LC7208 | China, Guangdong Province, Guangzhou city | bamboo | MW016500 | MW566367 | MW580540 | MW024528 | MW474486 | MW533819 | – | |
| LC7924 | China, Shandong Province | Capsicum sp. | MW016501 | MW566368 | MW580541 | MW024529 | MW474487 | MW533820 | – | |
| F. aquaticum | LC13615 | China, Guizhou Province, Zunyi city | water | MW016406 | MW566273 | MW580446 | MW024437 | MW474392 | MW533728 | – |
| LC13616 | China, Guizhou Province, Zunyi city | water | MW016407 | MW566274 | MW580447 | MW024438 | MW474393 | MW533729 | – | |
| CGMCC 3.20819 = LC7502 T | China, Guizhou Province, Zunyi city | water | MW016408 | MW566275 | MW580448 | MW024439 | MW474394 | MW533730 | – | |
| F. concentricum | LC1003 | China, Guangdong Province, Guangzhou city | Reineckia carnea | MW016409 | MW566276 | MW580449 | MW024440 | MW474395 | MW533731 | – |
| LC11489 = G1 | China, Beijing | Vitis sp. | MW016410 | MW566277 | MW580450 | MW024441 | MW474396 | MW533732 | – | |
| LC11491 = G5 | China, Beijing | Vitis sp. | MW016411 | MW566278 | MW580451 | MW024442 | MW474397 | MW533733 | – | |
| LC11507 = G36 | China, Beijing | Vitis sp. | MW016412 | MW566279 | MW580452 | MW024443 | MW474398 | MW533734 | – | |
| LC13617 = CQ1128 | China, Jiangsu Province, Changshu city | unknown plant | MW016413 | MW566280 | MW580453 | MW024444 | MW474399 | MW533735 | – | |
| LC13618 = F409 | Japan | Podocarpus macrophyllus | MW016414 | MW566281 | MW580454 | MW024445 | MW474400 | MW533736 | – | |
| LC13619 = FJWYS10-3 | China, Fujian Province, Wuyi Mountain | Musa nana | MW016415 | MW566282 | MW580455 | MW024446 | MZ399207 | MW533737 | – | |
| LC13620 = FJWYS3-1 | China, Fujian Province, Wuyi Mountain | Musa nana | MW016416 | MW566283 | MW580456 | MW024447 | MW474402 | MW533738 | – | |
| LC13621 = GXLB9-1-1 | China, Guangxi Zhuang Autonomous Region, Laibin city | Musa nana | MW016417 | MW566284 | MW580457 | MW024448 | MW474403 | MW533739 | – | |
| LC13623 = LJM1471 | China, Hainan Province, Haikou city | Maianthemum sp. | MW016419 | MW566286 | MW580459 | MW024450 | MW474405 | MW533741 | – | |
| LC13624 = M0514 | China, Fujian Province, Fuzhou city, Wuyi Mountain | Lablab sp. | MW016420 | MW566287 | MW580460 | MW024451 | MW474406 | MW533742 | – | |
| LC13647 = M0155 | China, Fujian Province, Fuzhou city | Lablab sp. | MW016457 | MW566324 | MW580497 | MW024485 | MW474443 | MW533776 | – | |
| LC13648 = M0155.2 | China, Fujian Province, Fuzhou city | Lablab sp. | MW016458 | MW566325 | MW580498 | MW024486 | MW474444 | MW533777 | – | |
| LC13649 = M0155.3 | China, Fujian Province, Fuzhou city | Lablab sp. | MW016459 | MW566326 | MW580499 | MW024487 | MW474445 | MW533778 | – | |
| LC4326 | China, Jiangxi Province | Aglaonema modestum | MW016421 | MW566288 | MW580461 | MW024452 | MW474407 | MW533743 | – | |
| LC4359 | China, Jiangxi Province | Hedera nepalensis | MW016422 | MW566289 | MW580462 | MW024453 | MW474408 | MW533744 | – | |
| LC7032 | China, Hainan Province | Musa nana | MW016423 | MW566290 | MW580463 | MW024454 | MW474409 | MW533745 | – | |
| F. elaeagni | CGMCC 3.20822 = LC13627 = CQ1053 T | China, Jiangsu Province, Suzhou city | Elaeagnus pungens | MW016426 | MW566293 | MW580466 | MW024457 | MW474412 | MW533748 | – |
| LC13628 = CQ1053.2 | China, Jiangsu Province, Suzhou city | Elaeagnus pungens | MW016427 | MW566294 | MW580467 | MW024458 | MW474413 | MW533749 | – | |
| LC13629 = CQ1053.3 | China, Jiangsu Province, Suzhou city | Elaeagnus pungens | MW016428 | MW566295 | MW580468 | MW024459 | MW474414 | MW533750 | – | |
| F. fujikuroi | LC13633 = F013 | USA | Glycine max | MW016432 | MW566299 | MW580472 | MW024460 | MW474418 | MW533751 | – |
| LC13634 = F032 | Japan | Acer palmatum | MW016433 | MW566300 | MW580473 | MW024461 | MW474419 | MW533752 | – | |
| LC13635 = F063 | USA | Sorghum bicolor | MW016434 | MW566301 | MW580474 | MW024462 | MW474420 | MW533753 | – | |
| LC13636 = F402 | Japan | Rhododendron simsii | MW016435 | MW566302 | MW580475 | MW024463 | MW474421 | MW533754 | – | |
| LC13637 = FJWYS2-1 | China, Fujian Province, Wuyi mountain | Musa nana | MW016436 | MW566303 | MW580476 | MW024464 | MW474422 | MW533755 | – | |
| LC13638 = GDQY3-1 | China, Guangdong Province, Qingyuan city | Musa nana | MW016437 | MW566304 | MW580477 | MW024465 | MW474423 | MW533756 | – | |
| LC13639 = GXBSNXS01-E1 | China, Guangxi Zhuang Autonomous Region, Baise city | Musa nana | MW016438 | MW566305 | MW580478 | MW024466 | MW474424 | MW533757 | – | |
| LC13640 = GXLZBDL06-E2 | China, Guangxi Zhuang Autonomous Region, Liuzhou city | Musa nana | MW016439 | MW566306 | MW580479 | MW024467 | MW474425 | MW533758 | – | |
| LC13641 = HBF4-8 | China, Hebei Province | Oryza sp. | MW016440 | MW566307 | MW580480 | MW024468 | MW474426 | MW533759 | – | |
| LC13642 = LJM1535 | China, Hainan Province, Wanning city | Panicum sp. | MW016441 | MW566308 | MW580481 | MW024469 | MW474427 | MW533760 | – | |
| LC13643 = LJM1536 | China, Hainan Province, Wanning city | Panicum sp. | MW016442 | MW566309 | MW580482 | MW024470 | MW474428 | MW533761 | – | |
| LC5916 | China, Jiangxi Province, Nanchang city | submerged wood | MW016443 | MW566310 | MW580483 | MW024471 | MW474429 | MW533762 | – | |
| LC5927 | China, Jiangxi Province, Nanchang city | submerged wood | MW016444 | MW566311 | MW580484 | MW024472 | MW474430 | MW533763 | – | |
| LC5945 | China, Jiangxi Province, Nanchang city | submerged wood | MW016445 | MW566312 | MW580485 | MW024473 | MW474431 | MW533764 | – | |
| LC5955 | China, Jiangxi Province, Nanchang city | submerged wood | MW016446 | MW566313 | MW580486 | MW024474 | MW474432 | MW533765 | – | |
| LC5979 | China, Jiangxi Province, Nanchang city | submerged wood | MW016447 | MW566314 | MW580487 | MW024475 | MW474433 | MW533766 | – | |
| LC6014 | China, Jiangxi Province, Nanchang city | submerged wood | MW016448 | MW566315 | MW580488 | MW024476 | MW474434 | MW533767 | – | |
| LC6015 | China, Jiangxi Province, Nanchang city | submerged wood | MW016449 | MW566316 | MW580489 | MW024477 | MW474435 | MW533768 | – | |
| LC6024 | China, Jiangxi Province, Nanchang city | submerged wood | MW016450 | MW566317 | MW580490 | MW024478 | MW474436 | MW533769 | – | |
| LC6973 | China, Jiangxi Province | Citrus reticulata | MW016451 | MW566318 | MW580491 | MW024479 | MW474437 | MW533770 | – | |
| LC7147 | China, Jiangxi Province | bamboo | MW016452 | MW566319 | MW580492 | MW024480 | MW474438 | MW533771 | – | |
| LC7864 | China, Guangxi Zhuang Autonomous Region | Poaceae sp. | MW016453 | MW566320 | MW580493 | MW024481 | MW474439 | MW533772 | – | |
| F. hechiense | CGMCC 3.20824 = LC13644 = GXHCSWL14-E1 T | China, Guangxi Zhuang Autonomous Region, Hechi city | Musa nana | MW016454 | MW566321 | MW580494 | MW024482 | MW474440 | MW533773 | – |
| LC13645 = GXHCSWL14-E12 | China, Guangxi Zhuang Autonomous Region, Hechi city, | Musa nana | MW016455 | MW566322 | MW580495 | MW024483 | MW474441 | MW533774 | – | |
| LC13646 = GXHCSWL14-E13 | China, Guangxi Zhuang Autonomous Region, Hechi city | Musa nana | MW016456 | MW566323 | MW580496 | MW024484 | MW474442 | MW533775 | – | |
| F. lumajangense | LC13650 = GXCZMQF02-1 | China, Guangxi Zhuang Autonomous Region, Chongzuo city | Musa nana | MW016461 | MW566328 | MW580501 | MW024489 | MW474447 | MW533780 | – |
| LC13651 = GXCZMQF02-2 | China, Guangxi Zhuang Autonomous Region, Chongzuo city | Musa nana | MW016462 | MW566329 | MW580502 | MW024490 | MW474448 | MW533781 | – | |
| LC13652 = MH0493 | China, Guangxi Zhuang Autonomous Region | Arenga caudata | MW016463 | MW566330 | MW580503 | MW024491 | MW474449 | MW533782 | – | |
| F. madaense | LC13614 = HBN5-22 | China, Hebei Province | Oryza sp. | MW016405 | MW566272 | MW580445 | MW024436 | MW474391 | MW533727 | – |
| F. mundagurra | LC13689 = LGS129 | China, Hainan Province | Paspalum vaginatum | MW016516 | MW566383 | MW580556 | MW024544 | MW474502 | MW533835 | – |
| LGS129.2 | China, Hainan Province | Paspalum vaginatum | MZ379241 | MZ399201 | MZ399211 | MZ399204 | MZ399208 | MZ399214 | – | |
| LGS129.3 | China, Hainan Province | Paspalum vaginatum | MZ379242 | MZ399202 | MZ399212 | MZ399205 | MZ399209 | MZ399215 | – | |
| F. panlongense | CGMCC 3.20825 = LC13656 = GXGLPLL15E2 T | China, Guangxi Zhuang Autonomous Region, Guilin city | Musa nana | MW016470 | MW566337 | MW580510 | MW024498 | MW474456 | MW533789 | – |
| F. proliferatum | F026 | China, Zhejiang Province, Ningbo city | Musa sp. | MZ379243 | MZ399203 | MZ399213 | MZ399206 | MZ399210 | MZ399216 | – |
| F. pseudocircinatum | LC13676 = F428 | China, Taiwan Province | Syzygium samarangense | MW016502 | MW566369 | MW580542 | MW024530 | MW474488 | MW533821 | – |
| LC13677 = F429 | China, Taiwan Province | Syzygium samarangense | MW016503 | MW566370 | MW580543 | MW024531 | MW474489 | MW533822 | – | |
| F. sacchari | LC1058 | China, Guangdong Province, Guangzhou city | Arundina graminifolia | MW016504 | MW566371 | MW580544 | MW024532 | MW474490 | MW533823 | – |
| LC13625 = F162 | Philippines | Musa sp. | MW016424 | MW566291 | MW580464 | MW024455 | MW474410 | MW533746 | – | |
| LC13626 = GDGZTHL40-E4 | China, Guangdong Province, Guangzhou city | Musa nana | MW016425 | MW566292 | MW580465 | MW024456 | MW474411 | MW533747 | – | |
| LC13657 = GXBSCGS01-E2 | China, Guangxi Zhuang Autonomous Region, Baise city | Musa nana | MW016471 | MW566338 | MW580511 | MW024499 | MW474457 | MW533790 | – | |
| LC13678 = GDGZ2-2 | China, Guangdong Province, Guangzhou city | Musa nana | MW016505 | MW566372 | MW580545 | MW024533 | MW474491 | MW533824 | – | |
| LC13679 = GXQZPSL01-E1 | China, Guangxi Zhuang Autonomous Region, Qinzhou city | Musa nana | MW016506 | MW566373 | MW580546 | MW024534 | MW474492 | MW533825 | – | |
| LC13680 = GXQZPSL01-E2 | China, Guangxi Zhuang Autonomous Region, Qinzhou city | Musa nana | MW016507 | MW566374 | MW580547 | MW024535 | MW474493 | MW533826 | – | |
| LC13681 = LJM1180 | China, Beijing | Poa annua | MW016508 | MW566375 | MW580548 | MW024536 | MW474494 | MW533827 | – | |
| F. subglutinans | LC13682 = F055 | USA | Glycine max | MW016509 | MW566376 | MW580549 | MW024537 | MW474495 | MW533828 | – |
| LC13683 = F057 | USA | Zea mays | MW016510 | MW566377 | MW580550 | MW024538 | MW474496 | MW533829 | – | |
| LC13684 = F154 | Canada | Glycine max | MW016511 | MW566378 | MW580551 | MW024539 | MW474497 | MW533830 | – | |
| LC13685 = F154-2 | Canada | Glycine max | MW016512 | MW566379 | MW580552 | MW024540 | MW474498 | MW533831 | – | |
| LC13686 = F154-3 | Canada | Glycine max | MW016513 | MW566380 | MW580553 | MW024541 | MW474499 | MW533832 | – | |
| F. temperatum | LC5848 | China, Guizhou Province | unidentified lichen | MW016460 | MW566327 | MW580500 | MW024488 | MW474446 | MW533779 | – |
| F. thapsinum | LC13687 = F103 | USA | Sorghum bicolor | MW016514 | MW566381 | MW580554 | MW024542 | MW474500 | MW533833 | – |
| LC13688 = F411 | USA | Glycine max | MW016515 | MW566382 | MW580555 | MW024543 | MW474501 | MW533834 | – | |
| F. verticillioides | LC13653 = F410 | Brazil | Glycine max | MW016464 | MW566331 | MW580504 | MW024492 | MW474450 | MW533783 | – |
| LC13654 = F412 | USA | Glycine max | MW016465 | MW566332 | MW580505 | MW024493 | MW474451 | MW533784 | – | |
| LC13655 = GDGZP4-1-1 | China, Guangdong Province, Guangzhou city | Musa nana | MW016466 | MW566333 | MW580506 | MW024494 | MW474452 | MW533785 | – | |
| LC2810 | China, Sichuan Province, Zhangjiajie | bamboo | MW016467 | MW566334 | MW580507 | MW024495 | MW474453 | MW533786 | – | |
| LC2818 | China, Beijing | Physosfegia virginiana | MW016468 | MW566335 | MW580508 | MW024496 | MW474454 | MW533787 | – | |
| LC5896 | China, Jiangxi Province, Nanchang city | submerged wood | MW016469 | MW566336 | MW580509 | MW024497 | MW474455 | MW533788 | – | |
| F. incarnatum-equiseti species complex | ||||||||||
| F. arcuatisporum | LC11639 = HA5-S04 | China, Hainan Province | Oryza sp. | MK280840 | MK289658 | MK289586 | MK289798 | MK289736 | MW533836 | – |
| CGMCC 3.19493 = LC12147 = LF1502 T | China, Hubei Province | Brassica campestris | MK280802 | MK289697 | MK289584 | MK289799 | MK289739 | MW533837 | – | |
| LC13690 = LGS034 | China, Beijing | soil | MW016517 | MW574182 | MW594360 | MW024545 | MW474503 | MW533838 | – | |
| LC13691 = LGS119 | China, Hainan Province | Paspalum vaginatum | MW016518 | MW574183 | MW594361 | MW024546 | MW474504 | MW533839 | – | |
| LC13692 = LJM0900 | China, Beijing | Poa annua | MW016519 | MW574184 | MW594362 | MW024547 | MW474505 | MW533840 | – | |
| LC13693 = LJM0939 | China, Beijing | unidentified grass | MW016520 | MW574185 | MW594363 | MW024548 | MW474506 | MW533841 | – | |
| LC13694 = LJM1441 | China, Hainan Province, Sanya city | Panicum sp. | MW016521 | MW574186 | MW594364 | MW024549 | MW474507 | MW533842 | – | |
| LC6026 | China, Jiangxi Province, Nanchang city | Nelumbo nucifera bloom | MK280792 | MK289667 | MK289585 | MK289800 | MK289770 | MW533843 | – | |
| F. citri | LC13695 = MH0430 | China, Guangxi Zhuang Autonomous Region | Castanopsis boisii | MW016522 | MW574187 | MW594365 | MW024550 | MW474508 | MW533844 | – |
| LC13696 = MH0439 | China, Guangxi Zhuang Autonomous Region | Castanopsis boisii | MW016523 | MW574188 | MW594366 | MW024551 | MW474509 | MW533845 | – | |
| LC13697 = MH0446 | China, Guangxi Zhuang Autonomous Region | Smilax corbularia | MW016524 | MW574189 | MW594367 | MW024552 | MW474510 | MW533846 | – | |
| LC13698 = YNTBL08E1 | China, Yunnan Province, Xishuangbanna | Musa nana | MW016525 | MW574190 | MW594368 | MW024553 | MW474511 | MW533847 | – | |
| LC4879 | China, Beijing | Amygdalus triloba | MK280820 | MK289665 | MK289615 | MK289827 | MK289768 | MW533848 | – | |
| CGMCC 3.19467 = LC6896 T | China, Hunan Province | Citrus reticulata | MK280803 | MK289668 | MK289617 | MK289828 | MK289771 | – | – | |
| LC7922 | China, Shandong Province | Capsicum sp. | MK280817 | MK289687 | MK289634 | MK289829 | MK289788 | – | – | |
| LC7937 | China, Shandong Province | Capsicum sp. | MK280797 | MK289693 | MK289640 | MK289830 | MK289794 | MW533849 | – | |
| F. compactum | LC13699 = LGS085 | China, Beijing | soil | MW016526 | MW574191 | MW594369 | MW024554 | MW474512 | – | – |
| LC13700 = LJM1181 | China, Beijing | Poa annua | MW016527 | MW574192 | MW594370 | MW024555 | MW474513 | MW533850 | – | |
| F. guilinense | CGMCC 3.19495 = LC12160 = GXGL9-3 T | China, Guangxi Zhuang Autonomous Region | Musa nana | MK280837 | MK289652 | MK289594 | MK289831 | MK289747 | MW533851 | – |
| F. hainanense | CGMCC 3.19478 = LC11638 = HA5-S03 T | China, Hainan Province | Oryza sp. | MK280836 | MK289657 | MK289581 | MK289833 | MK289735 | MW533852 | – |
| LC12161 = GXCZ-9-1 | China, Guangxi Zhuang Autonomous Region, Chongzuo city | Musa nana | MK280793 | MK289648 | MK289595 | MK289832 | MK289748 | MW533853 | – | |
| LC13701 = YNTBL31E2 | China, Yunnan Province, Xishuangbanna | Musa nana | MW016528 | MW574193 | MW594371 | MW024556 | MW474514 | MW533854 | – | |
| F. humuli | CQ1027 | China, Jiangsu Province | Ligustrum lucidum | MK280843 | MK289709 | MK289567 | MK289838 | MK289721 | MW533855 | – |
| CQ1032 | China, Jiangsu Province | Cedrela sp. | MK280844 | MK289710 | MK289568 | MK289839 | MK289722 | MW533856 | – | |
| CGMCC 3.19374 = CQ1039T | China, Jiangsu Province | Humulus scandens | MK280845 | MK289712 | MK289570 | MK289840 | MK289724 | MW533857 | – | |
| CQ1048 | China, Jiangsu Province | Viburnum sp. | MK280850 | MK289713 | MK289571 | MK289841 | MK289725 | MW533858 | – | |
| CQ1073 | China, Jiangsu Province | Liquidambar formosana | MK280848 | MK289714 | MK289572 | MK289842 | MK289726 | MW533859 | – | |
| CQ1133 | China, Jiangsu Province | Vinca major | MK280847 | MK289717 | MK289575 | MK289843 | MK289729 | MW533860 | – | |
| CQ969 | China, Jiangsu Province | Rosa sempervirens | MK280851 | MK289718 | MK289576 | MK289844 | MK289730 | MW533861 | – | |
| CQ970 | China, Jiangsu Province | Rosa sempervirens | MK280849 | MK289719 | MK289577 | MK289845 | MK289731 | MW533862 | – | |
| CQ975 | China, Jiangsu Province | Paederia foetida | MK280846 | MK289720 | MK289578 | MK289846 | MK289732 | MW533863 | – | |
| LC12158 = GDBYL14-E1 | China, Guangdong Province, Guangzhou city | Musa nana | MK280823 | MK289645 | MK289592 | MK289834 | MK289745 | MW533864 | – | |
| LC12159 = GDGZLHL14-E1 | China, Guangdong Province, Guangzhou city | Musa nana | MK280827 | MK289646 | MK289593 | MK289835 | MK289746 | MW533865 | – | |
| LC13702 = LJM1412 | China, Hainan Province, Haikou city | Megathyrsus sp. | MW016529 | MW574194 | MW594372 | MW024557 | MW474515 | MW533866 | – | |
| LC13703 = MH0134 | China, Guangxi Zhuang Autonomous Region | Coriaria nepalensis | MW016530 | MW574195 | MW594373 | MW024558 | MW474516 | MW533867 | – | |
| LC13704 = MH0240 | China, Guangxi Zhuang Autonomous Region | Chimonanthus praecox | MW016531 | MW574196 | MW594374 | MW024559 | MW474517 | MW533868 | – | |
| LC4490 | China, Jiangxi Province | Osmanthus sp. | MK280826 | MK289664 | MK289614 | MK289836 | MK289767 | MW533869 | – | |
| LC7003 | China, Hainan Province | Musa paradisiaca | MK280833 | MK289674 | MK289623 | MK289837 | MK289777 | MW533870 | – | |
| F. incarnatum | LC13705 = LGS051 | China, Beijing | soil | MW016532 | MW574197 | MW594375 | MW024560 | MW474518 | MW533871 | – |
| F. ipomoeae | CQ1099 | China, Jiangsu Province | Rhododendron pulchrum | MK280853 | MK289715 | MK289573 | MK289861 | MK289727 | MW533872 | – |
| CQ1132 | China, Jiangsu Province | Vinca major | MK280854 | MK289716 | MK289574 | MK289862 | MK289728 | MW533873 | – | |
| LC0166 | China, Beijing | Solanum lycopersicum | MK280780 | MK289659 | MK289579 | MK289848 | MK289733 | – | – | |
| LC0455 | China, Beijing | Hosta sp. | MK280819 | MK289660 | MK289580 | MK289849 | MK289734 | MW533874 | – | |
| LC12162 = GXLZCJL05-E2 | China, Guangxi Zhuang Autonomous Region, Liuzhou city | Musa nana | MK280795 | MK289655 | MK289596 | MK289847 | MK289749 | MW533875 | – | |
| LC12163 = M0027 | China, Fujian Province, Fuzhou city | Hibiscus syriacus | MK280790 | MK289700 | MK289597 | MK289857 | MK289750 | MW533876 | – | |
| LC12164 = M0028 | China, Fujian Province, Fuzhou city | Hibiscus syriacus | MK280822 | MK289701 | MK289598 | MK289858 | MK289751 | MW533877 | – | |
| CGMCC 3.19496 = LC12165 = M0111 T | China, Fujian Province | Ipomoea aquatica | MK280832 | MK289704 | MK289599 | MK289859 | MK289752 | MW533878 | – | |
| LC12166 = M0138 | China, Fujian Province, Fuzhou city | Lagenaria siceraria | MK280791 | MK289706 | MK289600 | MK289860 | MK289753 | – | – | |
| LC13706 = JXN4-3 | China, Jiangxi Province | Oryza sp. | MW016533 | MW574198 | MW594376 | MW024561 | MW474519 | – | – | |
| LC13707 = LGS036 | China, Beijing | soil | MW016534 | MW574199 | MW594377 | MW024562 | MW474520 | MW533879 | – | |
| LC13708 = LGS052 | China, Beijing | soil | MW016535 | MW574200 | MW594378 | MW024563 | MW474521 | – | – | |
| LC13709 = LGS071 | China, Beijing | soil | MW016536 | MW574201 | MW594379 | MW024564 | MW474522 | MW533880 | – | |
| LC13710 = LJM0958 | China, Beijing | Agrostis matsumurae | MW016537 | MW574202 | MW594380 | MW024565 | MW474523 | MW533881 | – | |
| LC5912 | China, Jiangxi Province | submerged wood | MK280821 | MK289666 | MK289616 | MK289850 | MK289769 | MW533882 | – | |
| LC6926 | China, Hubei Province | Oryza sativa | MK280799 | MK289670 | MK289619 | MK289851 | MK289773 | – | – | |
| LC7150 | China, Jiangxi Province | bamboo | MK280818 | MK289678 | MK289627 | MK289852 | MK289781 | MW533883 | – | |
| LC7923 | China, Shandong Province | Capsicum sp. | MK280800 | MK289688 | MK289635 | MK289853 | MK289789 | MW533884 | – | |
| LC7925 | China, Shandong Province | Capsicum sp. | MK280796 | MK289689 | MK289636 | MK289854 | MK289790 | – | – | |
| LC7936 | China, Shandong Province | Capsicum sp. | MK280785 | MK289692 | MK289639 | MK289855 | MK289793 | – | – | |
| LC7940 | China, Shandong province | Capsicum sp. | MK280798 | MK289695 | MK289642 | MK289856 | MK289796 | MW533885 | – | |
| F. irregulare | LC12145 | China, Guangdong Province | bamboo | MK280830 | MK289681 | MK289582 | MK289864 | MK289737 | – | – |
| LC12146 | China, Guangdong Province | bamboo | MK280831 | MK289682 | MK289583 | MK289865 | MK289738 | – | – | |
| LC13711 = LJM1544 | China, Hainan Province, Wanning city | Digitaria sp. | MW016538 | MW574203 | MW594381 | MW024566 | MW474524 | MW533886 | – | |
| LC13712 = LJM1545 | China, Hainan Province, Wanning city | Digitaria sp. | MW016539 | MW574204 | MW594382 | MW024567 | MW474525 | MW533887 | – | |
| LC13713 = MH0410 | China, Guangxi Zhuang Autonomous Region | Vigna unguiculata | MW016540 | MW574205 | MW594383 | MW024568 | MW474526 | MW533888 | – | |
| CGMCC 3.19489 = LC7188 T | China, Guangdong Province | bamboo | MK280829 | MK289680 | MK289629 | MK289863 | MK289783 | – | – | |
| F. lacertarum | LC7927 | China, Shandong Province | Capsicum sp. | MK280838 | MK289690 | MK289637 | MK289866 | MK289791 | – | – |
| LC7931 | China, Shandong Province | Capsicum sp. | MK280801 | MK289691 | MK289638 | MK289867 | MK289792 | – | – | |
| LC7942 | China, Shandong Province | Capsicum sp. | MK280834 | MK289696 | MK289643 | MK289868 | MK289797 | – | – | |
| F. luffae | CQ1038 | China, Jiangsu Province | Humulus scandens | MK280852 | MK289711 | MK289569 | MK289870 | MK289723 | MW533889 | – |
| CGMCC 3.19497 = LC12167 T | China, Fujian Province | Luffa aegyptiaca | MK280807 | MK289698 | MK289601 | MK289869 | MK289754 | – | – | |
| LC13714 = JXN4-19 | China, Jiangxi Province | Oryza sp. | MW016541 | MW574206 | MW594384 | MW024569 | MW474527 | – | – | |
| F. nanum | CGMCC 3.19498 = LC12168 = GXGL14-2 T | China, Guangxi Zhuang Autonomous Region | Musa nana | MK280794 | MK289651 | MK289602 | MK289871 | MK289755 | – | – |
| LC1384 | Saudi Arabia | Solanum lycopersicum | MK280842 | MK289661 | MK289611 | MK289872 | MK289764 | MW533890 | – | |
| LC1385 | Saudi Arabia | Solanum lycopersicum | MK280781 | MK289662 | MK289612 | MK289873 | MK289765 | MW533891 | – | |
| LC1516 | Saudi Arabia | Solanum lycopersicum | MK280782 | MK289663 | MK289613 | MK289874 | MK289766 | MW533892 | – | |
| F. pernambucanum | LC12148 = GDBYL12-E1 | China, Guangdong Province, Guangzhou city | Musa nana | MK280778 | MK289644 | MK289587 | MK289801 | MK289740 | MW533893 | – |
| LC12149 = GDGZP2-3 | China, Guangdong Province, Guangzhou city | Musa nana | MK280783 | MK289647 | MK289588 | MK289802 | MK289741 | MW533894 | – | |
| LC12151 = GXCZMQF01-3 | China, Guangxi Zhuang Autonomous Region, Chongzuo city | Musa nana | MK280825 | MK289649 | MK289589 | MK289803 | MK289742 | MW533895 | – | |
| LC12152 = GXCZMQF01-4 | China, Guangxi Zhuang Autonomous Region, Chongzuo city | Musa nana | MK280824 | MK289650 | MK289590 | MK289804 | MK289743 | MW533896 | – | |
| LC13715 = LJM1300 | China, Hainan Province, Haikou city | Heteropogon sp. | MW016542 | MW574207 | MW594385 | MW024570 | MW474528 | MW533897 | – | |
| LC13716 = LJM1312 | China, Hainan Province | Gerbera jamesonii | MW016543 | MW574208 | MW594386 | MW024571 | MW474529 | MW533898 | – | |
| LC13717 = LJM1438 | China, Hainan Province, Sanya city | Cyperus sp. | MW016544 | MW574209 | MW594387 | MW024572 | MW474530 | MW533899 | – | |
| LC13718 = LJM1523 | China, Hainan Province, Haikou city | Chamaedorea sp. | MW016545 | MW574210 | MW594388 | MW024573 | MW474531 | MW533900 | – | |
| LC13719 = LJM1529 | China, Hainan Province, Sanya city | Panicum sp. | MW016546 | MW574211 | MW594389 | MW024574 | MW474532 | MW533901 | – | |
| LC7014 | China, Hainan Province | Musa paradisiaca | MK280786 | MK289675 | MK289624 | MK289812 | MK289778 | – | – | |
| LC7019 | China, Hainan Province | Musa paradisiaca | MK280816 | MK289676 | MK289625 | MK289813 | MK289779 | – | – | |
| LC7040 | China, Hainan Province | Musa paradisiaca | MK280787 | MK289677 | MK289626 | MK289814 | MK289780 | MW533902 | – | |
| LC7157 | China, Jiangxi Province, Nanchang city | bamboo | MK280804 | MK289679 | MK289628 | MK289815 | MK289782 | – | – | |
| LC7842 | China, Hainan Province | Zea sp. | MK280813 | MK289684 | MK289631 | MK289817 | MK289785 | MW533903 | – | |
| LC7920 | China, Shandong Province | Capsicum sp. | MK280805 | MK289686 | MK289633 | MK289819 | MK289787 | – | – | |
| F. sulawesiense | LC12170 = GXNN-6 | China, Guangxi Zhuang Autonomous Region, Nanning city | Musa nana | MK280841 | MK289656 | MK289604 | MK289807 | MK289757 | MW533908 | – |
| LC12173 = M0010 | China, Fujian Province, Fuzhou city | Luffa aegyptiaca | MK280788 | MK289699 | MK289605 | MK289821 | MK289758 | – | – | |
| LC12174 = M0079 | China, Fujian Province, Fuzhou city | Ipomoea batatas | MK280815 | MK289702 | MK289606 | MK289822 | MK289759 | – | – | |
| LC12175 = M0110 | China, Fujian Province, Fuzhou city | Ipomoea aquatica | MK280808 | MK289703 | MK289607 | MK289823 | MK289760 | MW533909 | – | |
| LC12176 = M0117 | China, Fujian Province, Fuzhou city | Luffa aegyptiaca | MK280839 | MK289705 | MK289608 | MK289824 | MK289761 | – | – | |
| LC12177 = M0204 | China, Fujian Province, Fuzhou city | Colocasia esculenta | MK280809 | MK289707 | MK289609 | MK289825 | MK289762 | – | – | |
| LC12178 = M0751 | China, Fujian Province, Fuzhou city | Syngonium auritum | MK280789 | MK289708 | MK289610 | MK289826 | MK289763 | – | – | |
| LC13720 = JXN4-20 | China, Jiangxi Province | Oryza sp. | MW016547 | MW574212 | MW594390 | MW024575 | MW474533 | – | – | |
| LC13721 = MH0409 | China, Guangxi Zhuang Autonomous Region | Alocasia odora | MW016548 | MW574213 | MW594391 | MW024576 | MW474534 | MW533904 | – | |
| LC13722 = MH0423 | China, Guangxi Zhuang Autonomous Region | Acalypha insulana | MW016549 | MW574214 | MW594392 | MW024577 | MW474535 | MW533905 | – | |
| LC13723 = MH0447 | China, Guangxi Zhuang Autonomous Region | Smilax corbularia | MW016550 | MW574215 | MW594393 | MW024578 | MW474536 | MW533906 | – | |
| LC6897 | China, Hunan Province | Citrus reticulata | MK280810 | MK289669 | MK289618 | MK289808 | MK289772 | – | – | |
| LC6928 | China, Hubei Province | Oryza sativa | MK280835 | MK289671 | MK289620 | MK289809 | MK289774 | – | – | |
| LC6936 | China, Hubei Province | Oryza sativa | MK280835 | MK289620 | MK289671 | MK289809 | MK289774 | – | – | |
| LC6990 | China, Hainan Province | Musa paradisiaca | MK280814 | MK289673 | MK289622 | MK289811 | MK289776 | – | – | |
| LC7210 | China, Jiangxi Province, Nanchang city | bamboo | MK280804 | MK289679 | MK289628 | MK289815 | MK289782 | – | – | |
| LC7919 | China, Shandong Province | Capsicum sp. | MK280811 | MK289685 | MK289632 | MK289818 | MK289786 | – | – | |
| LC7939 | China, Shandong Province | Capsicum sp. | MK280806 | MK289694 | MK289641 | MK289820 | MK289795 | – | – | |
| F. tanahbumbuense | LC13724 = HBF4-12 | China, Hebei Province | Oryza sp. | MW016551 | MW574216 | MW594394 | MW024579 | MW474537 | – | – |
| LC13725 = HBN4-19 | China, Hebei Province | Oryza sp. | MW016552 | MW574217 | MW594395 | MW024580 | MW474538 | – | – | |
| LC13726 = LJM1369 | China, Hainan Province, Wanning city | Digitaria sp. | MW016553 | MW574218 | MW594396 | MW024581 | MW474539 | MW533910 | – | |
| F. lateritium species complex | ||||||||||
| F. cassiae | LC13727 = F092 | China, Yunnan Province | Coffea sp. | MW016554 | – | MW594307 | MW024582 | MW474540 | MW533911 | – |
| F. stilboides | LC13728 = CQ1109 | China, Jiangsu Province, Changshu city | Forsythia sp. | MW016555 | – | MW594308 | MW024583 | MW474541 | MW533912 | – |
| LC13729 = CQ993 | China, Jiangsu Province, Suzhou city | Hedera nepalensis var. sinensis | MW016556 | – | MW594309 | MW024584 | MW474542 | MW533913 | – | |
| LC13730 = F004 | Japan | Acer palmatum | MW016557 | – | MW594310 | MW024585 | MW474543 | MW533914 | – | |
| LC13731 = M0759 | China, Guangdong Province, Shenzhen city | Schima noronhae | MW016558 | – | MW594311 | MW024586 | MW474544 | MW533915 | – | |
| Fusarium sp. | LC13732 = F085 | China, Yunnan Province | Coffea sp. | MW016559 | – | MW594312 | MW024587 | MW474545 | MW533916 | – |
| LC13733 = F088 | China, Yunnan Province | Coffea sp. | MW016560 | – | MW594313 | MW024588 | MW474546 | MW533917 | – | |
| F. nisikadoi species complex | ||||||||||
| F. commune | LC11660 = HM259-R03 | China, Hainan Province | Oryza sp. | MW016699 | – | MW620160 | MW024727 | MW474685 | MW534045 | – |
| LC13823 = GDGZP2-2 | China, Guangdong Province, Guangzhou city | Musa nana | MW016700 | – | MW620161 | MW024728 | MW474686 | MW534046 | – | |
| LC13824 = GXQZPSRDE4 | China, Guangxi Zhuang Autonomous Region, Qinzhou city | Musa nana | MW016701 | – | MW620162 | MW024729 | MW474687 | – | – | |
| F. miscanthi | LC7503 | China, Guizhou Province, Zunyi city | water | MW016565 | – | MW594318 | MW024593 | MW474551 | MW533922 | – |
| F. paranisikadoi | CGMCC 3.20826 = LC2800 T | China, Beijing | unidentified grass | MW016561 | – | MW594314 | MW024589 | MW474547 | MW533918 | – |
| LC2819 | China, Beijing | unidentified grass | MW016562 | – | MW594315 | MW024590 | MW474548 | MW533919 | – | |
| LC2823 | China, Beijing | Pennisetum alopecuroides | MW016563 | – | MW594316 | MW024591 | MW474549 | MW533920 | – | |
| LC2824 | China, Beijing | unidentified grass | MW016564 | – | MW594317 | MW024592 | MW474550 | MW533921 | – | |
| F. oxysporum species complex | ||||||||||
| F. cugenangense | LC13734 = F001 | Japan | Acer palmatum | MW016566 | – | MW594319 | MW024594 | MW474552 | MW533923 | MW024379 |
| LC13735 = F080 | Brazil | Hordeum vulgare | MW016567 | – | MW594320 | MW024595 | MW474553 | MW533924 | MW024380 | |
| LC13736 = F422 | China, Zhejiang Province, Ningbo city | Solanum tuberosum | MW016568 | – | MW594321 | MW024596 | MW474554 | MW533925 | MW024381 | |
| LC4496 | China, Jiangxi Province | Smilax sp. | MW016571 | – | MW594324 | MW024599 | MW474557 | MW533928 | MW024384 | |
| F. curvatum | LC13739 = F155 | Netherlands | Tulipa gesneriana | MW016572 | – | MW594325 | MW024600 | MW474558 | MW533929 | MW024385 |
| F. duoseptatum | LC13740 = GXHCFoc1 | China, Guangxi Zhuang Autonomous Region, Hechi city | Musa nana | MW016573 | – | MW594326 | MW024601 | MW474559 | MW533930 | MW024386 |
| LC13741 = GXQZPSRDE3 | China, Guangxi Zhuang Autonomous Region, Qinzhou city | Musa nana | MW016574 | – | MW594327 | MW024602 | MW474560 | MW533931 | MW024387 | |
| F. elaeidis | LC13742 = M0765 | China, Guangdong Province, Shenzhen city | Caryota mitis | MW016575 | – | MW594328 | MW024603 | MW474561 | MW533932 | MW024388 |
| F. grosmichelii | GDGZP11-2-2 | China, Guangdong Province, Guangzhou city | Musa nana | OL744448 | – | OL771389 | OL771373 | OL771381 | OL771397 | OL780783 |
| GDZJLZ11-2-1 | China, Guangdong Province, Zhanjiang city | Musa nana | OL744449 | – | OL771390 | OL771374 | OL771382 | OL771398 | OL780784 | |
| GXCZMQS03E1 | China, Guangxi Zhuang Autonomous Region, Chongzuo city | Musa nana | OL744450 | – | OL771391 | OL771375 | OL771383 | OL771399 | OL780785 | |
| GXCZMQS03E2 | China, Guangxi Zhuang Autonomous Region, Chongzuo city | Musa nana | OL744451 | – | OL771392 | OL771376 | OL771384 | OL771400 | OL780786 | |
| JXF4-32 | China, Jiangxi Province | Oryza sp. | OL744452 | – | OL771393 | OL771377 | OL771385 | OL771401 | OL780787 | |
| JXF4-6 | China, Jiangxi Province | Oryza sp. | OL744453 | – | OL771394 | OL771378 | OL771386 | OL771402 | OL780788 | |
| JXN4-10 | China, Jiangxi Province | Oryza sp. | OL744454 | – | OL771395 | OL771379 | OL771387 | OL771403 | OL780789 | |
| M0676 | China, Guangdong Province, Shenzhen | Chamaerops humilis | OL744455 | – | OL771396 | OL771380 | OL771388 | OL771404 | OL780790 | |
| F. nirenbergiae | LC13752 = F014 | Italy | Hydrangea macrophylla | MW016585 | – | MW594338 | MW024613 | MW474571 | MW533942 | MW024398 |
| LC13753 = F051 | USA | Glycine max | MW016586 | – | MW594339 | MW024614 | MW474572 | MW533943 | MW024399 | |
| LC13754 = F077 | Italy | Olea europaea | MW016587 | – | MW594340 | MW024615 | MW474573 | MW533944 | MW024400 | |
| LC13755 = F153 | Canada | Glycine max | MW016588 | – | MW594341 | MW024616 | MW474574 | MW533945 | MW024401 | |
| LC13756 = F161 | Netherlands | Hippeastrum rutilum | MW016589 | – | MW594342 | MW024617 | MW474575 | MW533946 | MW024402 | |
| LC13757 = F418 | USA | Allium sativum | MW016590 | – | MW594343 | MW024618 | MW474576 | MW533947 | MW024403 | |
| LC13758 = GDZJLZ16-1 | China, Guangdong Province, Zhanjiang city | Musa nana | MW016591 | – | MW594344 | MW024619 | MW474577 | MW533948 | MW024404 | |
| LC13760 = M0579 | China, Guangdong Province | Caryota mitis | MW016593 | – | MW594346 | MW024621 | MW474579 | MW533950 | MW024406 | |
| LC2804 | China, Beijing | Setaria viridis | MW016594 | – | MW594347 | MW024622 | MW474580 | MW533951 | MW024407 | |
| F. odoratissimum | LC13761 = Foc4-6 | China, Guangxi Zhuang Autonomous Region | Musa sp. | MW016595 | – | MW594348 | MW024623 | MW474581 | MW533952 | MW024408 |
| LC13762 = Foc4-7 | China, Guangxi Zhuang Autonomous Region | Musa sp. | MW016596 | – | MW594349 | MW024624 | MW474582 | – | MW024409 | |
| LC13763 = Foc4-8 | China, Guangxi Zhuang Autonomous Region | Musa sp. | MW016597 | – | MW594350 | MW024625 | MW474583 | – | MW024410 | |
| LC13764 = GXLZCJL02-E5 | China, Guangxi Zhuang Autonomous Region, Liuzhou city | Musa nana | MW016598 | – | MW594351 | MW024626 | MW474584 | MW533953 | MW024411 | |
| LC13765 = GXNNS2 | China, Guangxi Zhuang Autonomous Region, Nanning city | Musa sp. | MW016599 | – | MW594352 | MW024627 | MW474585 | MW533954 | MW024412 | |
| F. oxysporum | LC13766 = F065 | China, Zhejiang Province, Ningbo city | Malus spectabilis | MW016600 | – | MW594353 | MW024628 | MW474586 | MW533955 | MW024413 |
| Fusarium sp. | LC13737 = J1BS1 | China, Shandong Province, Weifang city | Zingiber officinale | MW016569 | – | MW594322 | MW024597 | MW474555 | MW533926 | MW024382 |
| LC13743 = F163 | Brazil | Glycine max | MW016576 | – | MW594329 | MW024604 | MW474562 | MW533933 | MW024389 | |
| LC13744 = F416 | Brazil | Glycine max | MW016577 | – | MW594330 | MW024605 | MW474563 | MW533934 | MW024390 | |
| LC13745 = F151 | Australia | Hordeum vulgare | MW016578 | – | MW594331 | MW024606 | MW474564 | MW533935 | MW024391 | |
| LC13746 = F151-2 | Australia | Hordeum vulgare | MW016579 | – | MW594332 | MW024607 | MW474565 | MW533936 | MW024392 | |
| LC13747 = F151-3 | Australia | Hordeum vulgare | MW016580 | – | MW594333 | MW024608 | MW474566 | MW533937 | MW024393 | |
| LC13748 = F050 | Netherlands | Tulipa gesneriana | MW016581 | – | MW594334 | MW024609 | MW474567 | MW533938 | MW024394 | |
| LC13749 = F156 | Netherlands | Muscari botryoides | MW016582 | – | MW594335 | MW024610 | MW474568 | MW533939 | MW024395 | |
| LC13750 = GDZJLZ16-2 | China, Guangdong Province, Zhanjiang city | Musa nana | MW016583 | – | MW594336 | MW024611 | MW474569 | MW533940 | MW024396 | |
| LC13751 = GXCZ-4-1 | China, Guangxi Zhuang Autonomous Region, Chongzuo city | Musa nana | MW016584 | – | MW594337 | MW024612 | MW474570 | MW533941 | MW024397 | |
| LC13767 = LJM1259 | China, Guangxi Zhuang Autonomous Region | Passiflora edulis | MW016601 | – | MW594354 | MW024629 | MW474587 | MW533956 | MW024414 | |
| LC13768 = LJM1259-2 | China, Guangxi Zhuang Autonomous Region | Passiflora edulis | MW016602 | – | MW594355 | MW024630 | MW474588 | MW533957 | MW024415 | |
| LC13769 = LJM1259-3 | China, Guangxi Zhuang Autonomous Region | Passiflora edulis | MW016603 | – | MW594356 | MW024631 | MW474589 | MW533958 | MW024416 | |
| F. sambucinum species complex | ||||||||||
| F. acaciae-mearnsii | LC13786 = FJWYS2-3 | China, Fujian Province, Fuzhou city | Musa nana | MW016630 | – | MW620091 | MW024658 | MW474616 | MW533978 | – |
| F. armeniacum | LC2797 | China, Beijing | unidentified grass | MW016608 | – | MW620069 | MW024636 | MW474594 | MW533963 | – |
| LC2809 | China, Beijing | unidentified grass | MW016609 | – | MW620070 | MW024637 | MW474595 | MW533964 | – | |
| F. asiaticum | LC13773 = CQ974 | China, Jiangsu Province, Suzhou city | Paederia foetida | MW016607 | – | MW620068 | MW024635 | MW474593 | MW533962 | – |
| LC13774 = GXGLYSL08-1 | China, Guangxi Zhuang Autonomous Region, Guilin city | Musa nana | MW016610 | – | MW620071 | MW024638 | MW474596 | MW533965 | – | |
| LC13785 = F408 | China, Zhejiang Province, Ningbo city | Podocarpus macrophyllus | MW016629 | – | MW620090 | MW024657 | MW474615 | MW533977 | – | |
| LC13787 = GDBYL11-E1 | China, Guangdong Province, Guangzhou city | Musa nana | MW016631 | – | MW620092 | MW024659 | MW474617 | MW533979 | – | |
| LC13788 = GXGLPLL07E2 | China, Guangxi Zhuang Autonomous Region, Guilin city | Musa nana | MW016632 | – | MW620093 | MW024660 | MW474618 | MW533980 | – | |
| LC13789 = JXN5 | China, Jiangxi Province | Oryza sp. | MW016633 | – | MW620094 | MW024661 | MW474619 | MW533981 | – | |
| LC5153 | China, Jiangxi Province, Ganzhou city | Prunus persica | MW016635 | – | MW620096 | MW024663 | MW474621 | MW533982 | – | |
| LC5308 | China, Guizhou Province | air | MW016636 | – | MW620097 | MW024664 | MW474622 | MW533983 | – | |
| LC7143 | China, Jiangxi Province, Nanchang city | bamboo | MW016637 | – | MW620098 | MW024665 | MW474623 | MW533984 | – | |
| LC7494 | China, Guizhou Province, Zunyi city | carbonatite | MW016638 | – | MW620099 | MW024666 | MW474624 | MW533985 | – | |
| LC7495 | China, Guizhou Province, Zunyi city | carbonatite | MW016639 | – | MW620100 | MW024667 | MW474625 | MW533986 | – | |
| LC7500 | China, Guizhou Province, Zunyi city | carbonatite | MW016640 | – | MW620101 | MW024668 | MW474626 | MW533987 | – | |
| LC7501 | China, Guizhou Province, Zunyi city | soil | MW016641 | – | MW620102 | MW024669 | MW474627 | MW533988 | – | |
| F. graminearum | LC13775 = F056 | USA | Zea mays | MW016611 | – | MW620072 | MW024639 | MW474597 | MW533966 | – |
| LC13776 = F110 | Italy | Pennisetum orientale | MW016612 | – | MW620073 | MW024640 | MW474598 | – | – | |
| F. kyushuense | LC0725 | China, Zhejiang Province, Quzhou city | Chamaedaphne calyculata | MW016613 | – | MW620074 | MW024641 | MW474599 | MW533967 | – |
| LC1114 | China | Lithocarpus glabra | MW016614 | – | MW620075 | MW024642 | MW474600 | MW533968 | – | |
| LC13777 = F179 | Philippines | unidentified plant | MW016615 | – | MW620076 | MW024643 | MW474601 | MW533969 | – | |
| LC13778 = GXLZ6-1 | China, Guangxi Zhuang Autonomous Region, Liuzhou city | Musa nana | MW016616 | – | MW620077 | MW024644 | MW474602 | MW533970 | – | |
| LC5936 | China, Jiangxi Province, Nanchang city | submerged wood | MW016617 | – | MW620078 | MW024645 | MW474603 | MW533971 | – | |
| LC7000 | China, Hainan Province | Musa paradisiaca | MW016618 | – | MW620079 | MW024646 | MW474604 | MW533972 | – | |
| F. longipes | LC13779 = LGS185 | China, Hainan Province | Paspalum vaginatum | MW016619 | – | MW620080 | MW024647 | MW474605 | – | – |
| F. meridionale | LC13780 = F087 | China, Yunnan Province | Coffea sp. | MW016620 | – | MW620081 | MW024648 | MW474606 | – | – |
| LC7067 | China, Yunnan Province | Musa basjoo | MW016621 | – | MW620082 | MW024649 | MW474607 | MW533973 | – | |
| LC7496 | China, Guizhou Province, Zunyi city | carbonatite | MW016622 | – | MW620083 | MW024650 | MW474608 | MW533974 | – | |
| F. nepalense | LC13781 = GDBYL14-E3 | China, Guangdong Province, Guangzhou city | Musa nana | MW016623 | – | MW620084 | MW024651 | MW474609 | MW533975 | – |
| LC13782 = GXGLPLL14-1 | China, Guangxi Zhuang Autonomous Region, Guilin city | Musa nana | MW016624 | – | MW620085 | MW024652 | MW474610 | – | – | |
| LC6678 | China, Yunnan Province, Xishuangbanna | Camellia sinensis | MW016625 | – | MW620086 | MW024653 | MW474611 | MW533976 | – | |
| F. poae | LC13783 = F150 | Canada | Hordeum vulgare | MW016626 | – | MW620087 | MW024654 | MW474612 | – | – |
| LC6917 | China, Hubei Province | Oryza sativa | MW016627 | – | MW620088 | MW024655 | MW474613 | – | – | |
| F. transvaalense | LC13784 = F157 | USA | Medicago sativa | MW016628 | – | MW620089 | MW024656 | MW474614 | – | – |
| F. ussurianum | LC13790 = LJM1343 | China, Hainan Province, Wanning city | Rhynchospora sp. | MW016634 | – | MW620095 | MW024662 | MW474620 | – | – |
| LC7573 | China, Tibet Autonomous Region | Poaceae sp. | MW016642 | – | MW620103 | MW024670 | MW474628 | – | – | |
| LC7574 | China, Tibet Autonomous Region | Poaceae sp. | MW016643 | – | MW620104 | MW024671 | MW474629 | MW533989 | – | |
| F. tricinctum species complex | ||||||||||
| F. acuminatum | LC13791 = F034 | Argentina | Glycine max | MW016644 | – | MW620105 | MW024672 | MW474630 | MW533990 | – |
| LC13794 = F111 | Italy | Feijoa sellowiana | MW016647 | – | MW620108 | MW024675 | MW474633 | MW533993 | – | |
| LC13795 = GM18 | China, Qinghai Province | Hylotelephium erythrostictum | MW016648 | – | MW620109 | MW024676 | MW474634 | MW533994 | – | |
| LC13796 = GM80 | China, Qinghai Province | Hylotelephium erythrostictum | MW016649 | – | MW620110 | MW024677 | MW474635 | MW533995 | – | |
| LC13797 = LF1633 | China, Hubei Province, Xiangyang city | Brassica sp. | MW016650 | – | MW620111 | MW024678 | MW474636 | MW533996 | – | |
| LC13798 = LF1636 | China, Hubei Province, Xiangyang city | Brassica sp. | MW016651 | – | MW620112 | MW024679 | MW474637 | MW533997 | – | |
| LC13799 = LGS021 | China, Beijing | soil | MW016652 | – | MW620113 | MW024680 | MW474638 | MW533998 | – | |
| LC5227 | China, Neimenggu Province, Huhehaote city | Prunus sp. | MW016654 | – | MW620115 | MW024682 | MW474640 | MW534000 | – | |
| F. alpinum | LC2853 | China, Yunnan Province | unidentified plant | MW016684 | – | MW620145 | MW024712 | MW474670 | MW534030 | – |
| LC2854 | China, Yunnan Province | unidentified plant | MW016685 | – | MW620146 | MW024713 | MW474671 | MW534031 | – | |
| LC6034 | China, Tibet Autonomous Region | Fabaceae sp. | MW016686 | – | MW620147 | MW024714 | MW474672 | MW534032 | – | |
| LC6037 | China, Tibet Autonomous Region | Fabaceae sp. | MW016687 | – | MW620148 | MW024715 | MW474673 | MW534033 | – | |
| LC6043 | China, Tibet Autonomous Region | Fabaceae sp. | MW016688 | – | MW620149 | MW024716 | MW474674 | MW534034 | – | |
| CGMCC 3.20818 = LC6045 T | China, Tibet Autonomous Region | Fabaceae sp. | MW016689 | – | MW620150 | MW024717 | MW474675 | MW534035 | – | |
| F. avenaceum | LC13801 = F010 | Italy | Hydrangea macrophylla | MW016655 | – | MW620116 | MW024683 | MW474641 | MW534001 | – |
| LC13802 = F038 | Australia | Trifolium repens | MW016656 | – | MW620117 | MW024684 | MW474642 | MW534002 | – | |
| LC13803 = F039 | Australia | Trifolium repens | MW016657 | – | MW620118 | MW024685 | MW474643 | MW534003 | – | |
| LC13804 = F071 | USA | Acer saccharum | MW016658 | – | MW620119 | MW024686 | MW474644 | MW534004 | – | |
| LC13805 = F403 | USA | Acer truncatum | MW016659 | – | MW620120 | MW024687 | MW474645 | MW534005 | – | |
| LC13806 = F404 | USA | Acer truncatum | MW016660 | – | MW620121 | MW024688 | MW474646 | MW534006 | – | |
| LC13808 = GM149 | China, Qinghai Province | Halenia sibirica | MW016662 | – | MW620123 | MW024690 | MW474648 | MW534008 | – | |
| LC13809 = GM30 | China, Qinghai Province | Bidens bipinnata | MW016663 | – | MW620124 | MW024691 | MW474649 | MW534009 | – | |
| LC13811 = GM71 | China, Qinghai Province | Halenia sibirica | MW016665 | – | MW620126 | MW024693 | MW474651 | MW534011 | – | |
| LC6044 | China, Tibet Autonomous Region | Fabaceae sp. | MW016667 | – | MW620128 | MW024695 | MW474653 | MW534013 | – | |
| LC6321 | China, Guizhou Province | Camellia sinensis | MW016668 | – | MW620129 | MW024696 | MW474654 | MW534014 | – | |
| LC6328 | China, Guizhou Province | Camellia sinensis | MW016669 | – | MW620130 | MW024697 | MW474655 | MW534015 | – | |
| LC6376 | China, Guizhou Province | Camellia sinensis | MW016670 | – | MW620131 | MW024698 | MW474656 | MW534016 | – | |
| LC6387 | China, Guizhou Province | Camellia sinensis | MW016671 | – | MW620132 | MW024699 | MW474657 | MW534017 | – | |
| LC6388 | China, Guizhou Province | Camellia sinensis | MW016672 | – | MW620133 | MW024700 | MW474658 | MW534018 | – | |
| LC6389 | China, Guizhou Province | Camellia sinensis | MW016673 | – | MW620134 | MW024701 | MW474659 | MW534019 | – | |
| LC7584 | China, Tibet Autonomous Region | Poaceae sp. | MW016674 | – | MW620135 | MW024702 | MW474660 | MW534020 | – | |
| F. chongqingense | LC13813 | China, Chongqing | Bothrocaryum controversum | MW016675 | – | MW620136 | MW024703 | MW474661 | MW534021 | – |
| LC13814 | China, Chongqing | Bothrocaryum controversum | MW016676 | – | MW620137 | MW024704 | MW474662 | MW534022 | – | |
| CGMCC 3.20821 = LC4957 T | China, Chongqing | Bothrocaryum controversum | MW016677 | – | MW620138 | MW024705 | MW474663 | MW534023 | – | |
| F. iranicum | LC1112 | China | Lithocarpus glabra | MW016678 | – | MW620139 | MW024706 | MW474664 | MW534024 | – |
| F. paeoniae | LC13807 = GM123 | China, Qinghai Province | Plantago sp. | MW016661 | – | MW620122 | MW024689 | MW474647 | MW534007 | – |
| LC13810 = GM65 | China, Qinghai Province | Gentiana scabra | MW016664 | – | MW620125 | MW024692 | MW474650 | MW534010 | – | |
| LC13812 = GM85 | China, Qinghai Province | Gentiana scabra | MW016666 | – | MW620127 | MW024694 | MW474652 | MW534012 | – | |
| LC13815 = GM56 | China, Qinghai Province | Elymus dahuricus | MW016679 | – | MW620140 | MW024707 | MW474665 | MW534025 | – | |
| LC13816 = YZG10-2 | China, Qinghai Province | Populus sp. | MW016680 | – | MW620141 | MW024708 | MW474666 | MW534026 | – | |
| CGMCC 3.20817 = LC13817 = YZG12-2 T | China, Qinghai Province | Paeonia lactiflora | MW016681 | – | MW620142 | MW024709 | MW474667 | MW534027 | – | |
| LC5166 | China, Qinghai Province | Crataegus monogyna | MW016682 | – | MW620143 | MW024710 | MW474668 | MW534028 | – | |
| LC7358 | China, Tibet Autonomous Region | Poaceae sp. | MW016683 | – | MW620144 | MW024711 | MW474669 | MW534029 | – | |
| F. tricinctum | LC0453 | China, Beijing | Hosta sp. | MW016690 | – | MW620151 | MW024718 | MW474676 | MW534036 | – |
| LC0459 | China, Beijing | Zamia pumila | MW016691 | – | MW620152 | MW024719 | MW474677 | MW534037 | – | |
| LC13818 = F005 | Japan | Acer palmatum | MW016692 | – | MW620153 | MW024720 | MW474678 | MW534038 | – | |
| LC13819 = F020 | Poland | Clematis sp. | MW016693 | – | MW620154 | MW024721 | MW474679 | MW534039 | – | |
| LC13820 = F033 | Japan | Acer palmatum | MW016694 | – | MW620155 | MW024722 | MW474680 | MW534040 | – | |
| LC13821 = F400 | Japan | Chaenomeles japonica | MW016695 | – | MW620156 | MW024723 | MW474681 | MW534041 | – | |
| LC13822 = PH53 | China, Zhejiang Province, Ningbo city | unidentified plant | MW016696 | – | MW620157 | MW024724 | MW474682 | MW534042 | – | |
| LC5032 | China, Jiangxi Province, Ganzhou city | Litsea sp. | MW016697 | – | MW620158 | MW024725 | MW474683 | MW534043 | – | |
| LC5034 | China, Jiangxi Province, Ganzhou city | Litsea sp. | MW016698 | – | MW620159 | MW024726 | MW474684 | MW534044 | – | |
| Neocosmospora | ||||||||||
| N. brevis | LC2116 | China, Jiangxi Province, Ganzhou city | submerged wood | MW016702 | – | MW620163 | MW024730 | MW474688 | – | – |
| N. diminuta | LC13825 = F009 | Japan | Acer palmatum | MW016703 | – | MW620164 | MW024731 | MW474689 | MW534047 | – |
| N. falciformis | LC11569 = G649 | China | Vitis sp. | MW016704 | – | MW620165 | MW024732 | MW474690 | MW534048 | – |
| LC11572 = G694 | China | Vitis sp. | MW016705 | – | MW620166 | MW024733 | MW474691 | MW534049 | – | |
| LC13826 = LGS175 | China, Hainan Province | Paspalum vaginatum | MW016706 | – | MW620167 | MW024734 | MW474692 | – | – | |
| LC13827 = LGS230 | China, Hainan Province | Paspalum vaginatum | MW016707 | – | MW620168 | MW024735 | MW474693 | – | – | |
| LC13828 = LJM1271 | China, Guangxi Zhuang Autonomous Region | Passiflora edulis | MW016708 | – | MW620169 | MW024736 | MW474694 | MW534050 | – | |
| LC13829 = LJM1289 | China, Hainan Province | Paspalum vaginatum | MW016709 | – | MW620170 | MW024737 | MW474695 | – | – | |
| LC13830 = LJM1295 | China, Hainan Province | Paspalum vaginatum | MW016710 | – | MW620171 | MW024738 | MW474696 | – | – | |
| N. lithocarpi | CGMCC 3.20827 = LC1113 T | China | Lithocarpus glabra | MW016711 | – | MW620172 | MW024739 | MW474697 | MW534051 | – |
| LC13831 | China | Lithocarpus glabra | MW016712 | – | MW620173 | MW024740 | MW474698 | MW534052 | – | |
| LC13832 | China | Lithocarpus glabra | MW016713 | – | MW620174 | MW024741 | MW474699 | MW534053 | – | |
| N. longissima | LC13833 = F301 | Japan | Armeniaca mume | MW016714 | – | MW620175 | MW024742 | MW474700 | MW534054 | – |
| LC13834 = F303 | Japan | Armeniaca mume | MW016715 | – | MW620176 | MW024743 | MW474701 | MW534055 | – | |
| N. metavorans | LC5930 | China, Jiangxi Province, Nanchang city | submerged wood | MW016716 | – | MW620177 | MW024744 | MW474702 | MW534056 | – |
| LC5933 | China, Jiangxi Province, Nanchang city | submerged wood | MW016717 | – | MW620178 | MW024745 | MW474703 | MW534057 | – | |
| N. oblonga | LC7499 | China, Guizhou Province, Zunyi city | carbonatite | MW016718 | – | MW620179 | MW024746 | MW474704 | MW534058 | – |
| N. paraeumartii | LC13835 = F066 | Japan | Acer sp. | MW016719 | – | MW620180 | MW024747 | MW474705 | MW534059 | – |
| LC13836 = M0478 | China, Fujian Province, Fuzhou city | Castanopsis fargesii | MW016720 | – | MW620181 | MW024748 | MW474706 | MW534060 | – | |
| N. petroliphila | LC1120 | China | Lithocarpus glabra | MW016721 | – | MW620182 | MW024749 | MW474707 | – | – |
| N. pisi | LC13837 = F073 | USA | Acer platanoides | MW016722 | – | MW620183 | MW024750 | MW474708 | – | – |
| N. pseudensiformis | LC13838 = LJM1257 | China, Guangxi Zhuang Autonomous Region | Passiflora edulis | MW016723 | – | MW620184 | MW024751 | MW474709 | MW534061 | – |
| LC13839 = LJM1263 | China, Guangxi Zhuang Autonomous Region | Passiflora edulis | MW016724 | – | MW620185 | MW024752 | MW474710 | MW534062 | – | |
| LC13840 = LJM1273 | China, Guangxi Zhuang Autonomous Region | Passiflora edulis | MW016725 | – | MW620186 | MW024753 | MW474711 | MW534063 | – | |
| N. silvicola | LC5482 | China, Guizhou Province | faeces | MW016726 | – | MW620187 | MW024756 | MW474712 | MW534066 | – |
| N. solani | LC13841 = 6S1 | China, Shandong Province, Weifang city | Capsicum annuum | MW016727 | – | MW620188 | MW024757 | MW474713 | MW534067 | – |
| LC13842 = F002 | Japan | Acer palmatum | MW016728 | – | MW620189 | MW024758 | MW474714 | MW534068 | – | |
| LC13843 = F016 | Italy | Syringa vulgaris | MW016729 | – | MW620190 | MW024759 | MW474715 | MW534069 | – | |
| LC13844 = HBN6-5 | China, Hebei Province | Oryza sp. | MW016730 | – | MW620191 | MW024760 | MW474716 | – | – | |
| LC13845 = J3R1 | China, Shandong Province, Weifang city | Zingiber officinale | MW016731 | – | MW620192 | MW024761 | MW474717 | MW534070 | – | |
| LC13846 = J3R2 | China, Shandong Province, Weifang city | Zingiber officinale | MW016732 | – | MW620193 | MW024762 | MW474718 | MW534071 | – | |
| LC13847 = LGS032 | China, Beijing | soil | MW016733 | – | MW620194 | MW024763 | MW474719 | MW534072 | – | |
| LC13848 = LGS033 | China, Beijing | soil | MW016734 | – | MW620195 | MW024764 | MW474720 | – | – | |
| LC13849 = LGS054 | China, Beijing | soil | MW016735 | – | MW620196 | MW024765 | MW474721 | MW534073 | – | |
| LC3717 | China, Guangxi Zhuang Autonomous Region, Nanning city | soil | MW016736 | – | MW620197 | MW024766 | MW474722 | MW534074 | – | |
| LC3785 | China, Shanxi Province, Baode city | soil | MW016737 | – | MW620198 | MW024767 | MW474723 | MW534075 | – | |
| LC3932 | China, Shanxi Province, Baode city | compost | MW016738 | – | MW620199 | MW024768 | MW474724 | MW534076 | – | |
| LC5548 | China, Guizhou Province | soil | MW016739 | – | MW620200 | MW024769 | MW474725 | – | – | |
| N. stercicola | LC5387 | China, Guizhou Province | soil | MW016740 | – | MW620201 | MW024770 | MW474726 | MW534077 | – |
Note: T = Ex-type specimen of new species; A. = Albonectria, B. = Bisifusarium, F. = Fusarium, N. = Neocosmospora.
Morphological observation
Examined isolates were incubated on synthetic nutrient poor agar plates (SNA; Nirenberg 1976) for 7 d at 25 °C. Agar pieces of approximately 5 × 5 mm were cut from the edge of colonies and transferred onto media for morphological characterisation. Culture characteristics, including colony morphology, pigmentation and odour, were observed after 7 d incubation in the dark on PDA, oatmeal agar (OA; Crous et al. 2019), and SNA. Colours were rated according to the colour charts of Kornerup & Wanscher (1978). Sporodochia were induced by incubating under a 12/12 h near-ultraviolet light/dark cycle, on SNA and water agar amended with sterilised pieces of carnation leaves (CLA; Snyder & Hansen 1947, Fisher et al. 1982) at 25 °C, respectively. Micromorphological characteristics were examined and photo-documented with water as mounting medium under a Nikon 80i microscope with Differential Interference Contrast (DIC) optics, and a Nikon SMZ1500 dissecting microscope. For each species, respectively 30 conidiophores, conidiogenous cells and chlamydospores, 50 micro- and macroconidia were mounted and randomly measured to calculate the mean size and standard deviation (SD).
DNA extraction and amplification
Genomic DNA was extracted from fungal mycelia grown on PDA, using a modified CTAB protocol as described in Guo et al. (2000). Seven loci, including the 5.8S nuclear ribosomal RNA gene with the two flanking internal transcribed spacer (ITS) re gions, intergenic spacer region of the rDNA (IGS), partial trans lation elongation factor (tef1), partial calmodulin (cam), partial RNA polymerase largest subunit (rpb1), partial RNA polymerase second largest subunit (rpb2) gene regions, and partial β-tubulin (tub2), were amplified and sequenced, respectively. The primer pairs and PCR amplification procedures following protocols described by O’Donnell et al. (1998a, b, 2008, 2009a, b, 2010), Crous et al. (2009, 2021), and Lombard et al. (2015), are listed in Table 2. PCR amplifications were performed in a reaction mixture consisting of 12.5 μL 2 × Taq PCR Master Mix (Vazyme Biotech Co., Ltd, Nanjing, China), 1 μL each of 10 μM primers, 1 μL of the undiluted genomic DNA, adjusted to a final volume of 25 μL with distilled deionized water. The PCR products were visualised on 1 % agarose electrophoresis gel. Sequencing was done bi-directionally, conducted by the Tianyi Huiyuan Company (Beijing, China). Consensus sequences were obtained using SeqMan of the Lasergene software package v. 14.1 (DNAstar, Madison, Wisconsin, USA).
Table 2.
rimers information of PCR amplification of the seven loci.
| Locus | Primer | Sequence of Primer (5’-3’) | Annealing temperature (°C) | References |
|---|---|---|---|---|
| ITS | ITS5 | GGAAGTAAAAGTCGTAACAAGG | 55 | White et al. (1990) |
| ITS4 | TCCTCCGCTTATTGATATGC | |||
| IGS | iNL11 | AGGCTTCGGCTTAGCGTCTTAG | 55 | O’Donnell et al. (2009a) |
| iCNS1 | TTTCGCAGTGAGGTCGGCAG | |||
| tef1 | EF1 | ATGGGTAAGGARGACAAGAC | 55 | O’Donnell et al. (1998b) |
| EF2 | GGARGTACCAGTSATCATG | |||
| cam | CL1 | GARTWCAAGGAGGCCTTCTC | 55 | O’Donnell et al. (2000a) |
| CL2A | TTTTTGCATCATGAGTTGGAC | |||
| rpb1 | RPB1-Fa | CAYAARGARTCYATGATGGGWC | 58 (5 cycles)?57 (5)?56 (35) | O’Donnell et al. (2010) |
| RPB1-G2R | GTCATYTGDGTDGCDGGYTCDCC | |||
| rpb2 | RPB2-5f2 | GGGGWGAYCAGAAGAAGGC | 57 | Reeb et al. (2004) |
| RPB2-11ar | GCRTGGATCTTRTCRTCSACC | Liu et al. (1999) | ||
| tub2 | T1 | AACATGCGTGAGATTGTAAGT | 54 | O’Donnell & Cigelnik (1997) |
| T2 | TAGTGACCCTTGGCCCAGTTG |
Phylogenetic analyses
Sequences of the 425 fusarioid strains studied in this study (356 from China, 69 intercepted from 13 other countries) are listed in Table 1. For each locus, sequences were aligned us ing MAFFT v. 7 (Katoh et al. 2017), and the alignments were manually adjusted where necessary. The best-fit nucleotide substitution models under the Akaike Information Criterion (AIC) were selected using jModelTest v. 2.1.7 (Posada 2008, Darriba et al. 2012). Alignments derived from this study were deposited in TreeBASE (submission ID 29103), taxonomic novelties in MycoBank, and new sequences in NCBIs GenBank database (https://www.ncbi.nlm.nih.gov/; assession numbers shown in Table 1). Phylogenetic analyses of both individual and combined datasets were performed using Bayesian inference (BI) and Maximum- likelihood (ML) methods. The BI analyses were conducted using MrBayes v. 3.2.1 (Huelsenbeck & Ronquist 2001) following the protocol of Wang et al. (2019), with optimisation of each locus treated as partitions in combined analyses, based on the Markov Chain Monte Carlo (MCMC) approach (Ronquist et al. 2012). All characters were equally weighted, and gaps were treated as missing data. Stationarity of analysis was determined by examining the standard deviation of split frequencies (< 0.01) and –ln likelihood plots in AWTY (Nylander et al. 2008). The ML analyses were conducted using PhyML v. 3.0 (Guindon et al. 2010), with 1000 bootstrap replicates. The general time reversible model was applied with an invariable gamma-distributed rate variation (GTR+I+G).
RESULTS
Phylogenetic analyses
Analyses of the generic level phylogeny of fusarioid fungi were conducted by using a combined tef1, rpb1, and rpb2 dataset that included 643 bp for tef1, 1583 bp of rpb1, and 1311 bp for rpb2. For the BI and ML analyses, a GTR+I+G model was selected for the combined tef1-rpb1-rpb2 dataset. The combined tef1, rpb1, and rpb2 phylogeny (Fig. 1) revealed that the Chinese isolates clustered into nine species complexes in Fusarium, and two allied genera (Bisifusarium and Neocosmospora). Isolate LC13606 from Podocarpus macrophyllus imported from Japan was closest to Albonectria rigidiuscula CBS 122570 (Fig. 1).
Fig. 1.
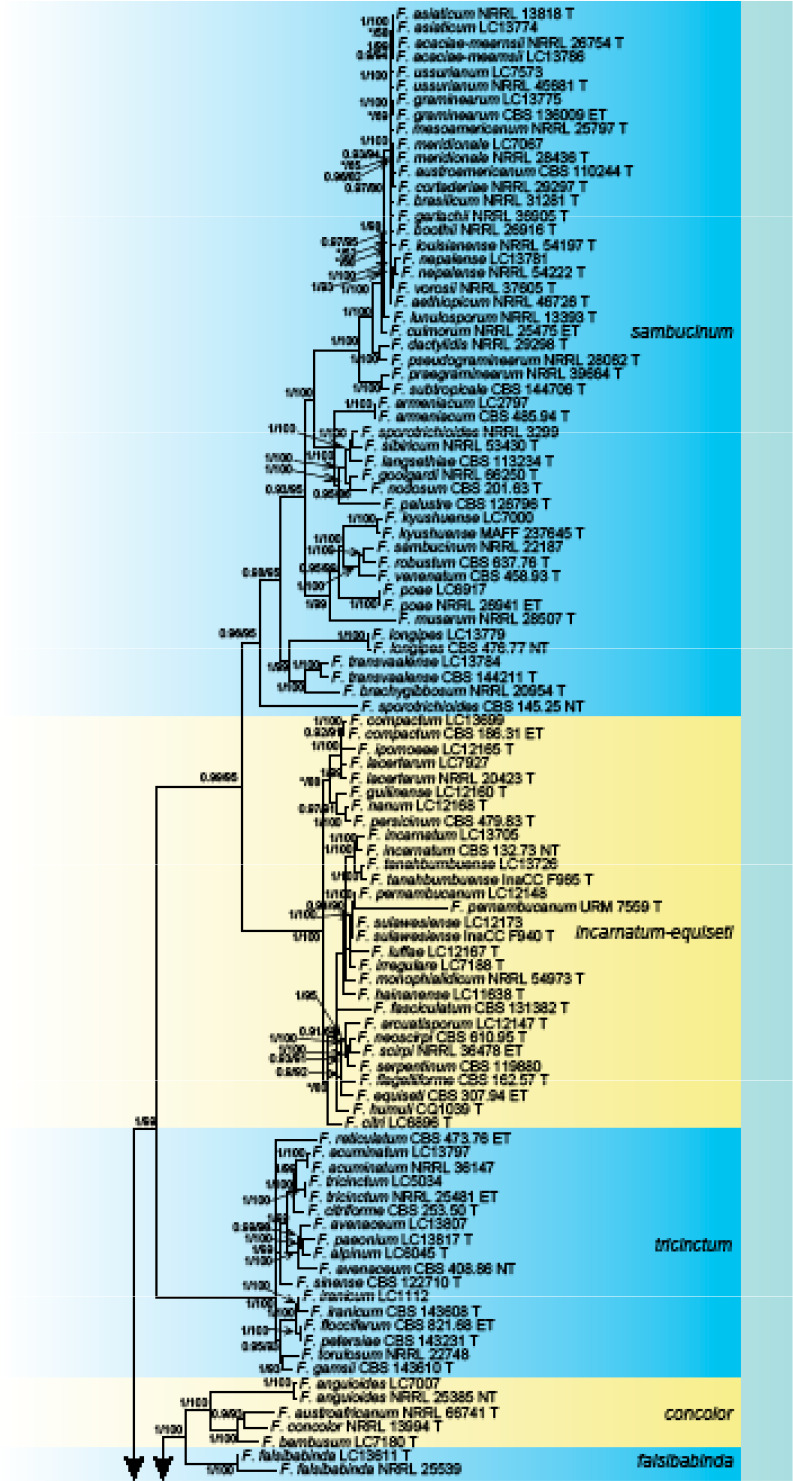

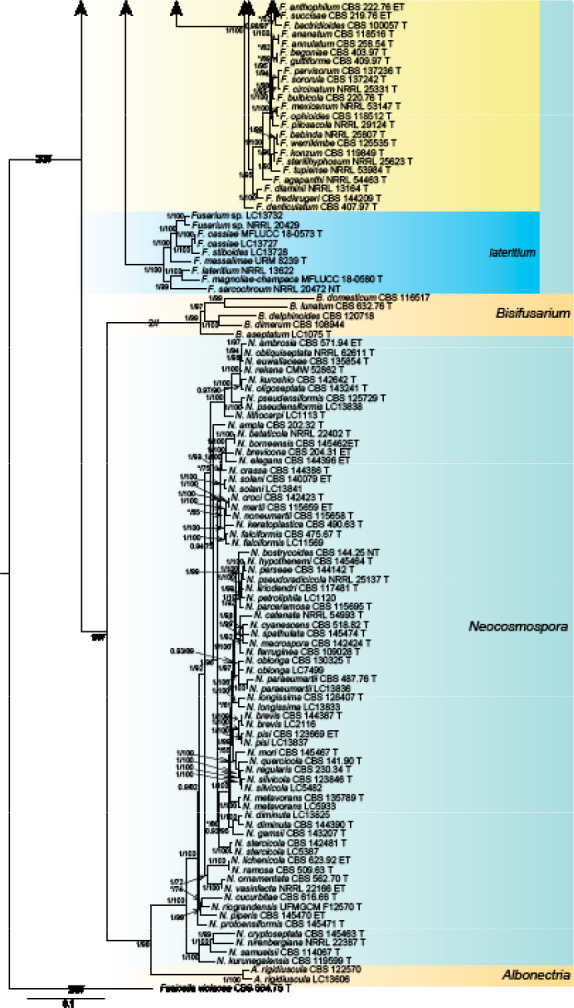
Fifty percent majority rule consensus tree from a Bayesian analysis based on a three-locus combined dataset (tef1, rpb1, and rpb2) showing the phylogenetic relationships of Fusarium and allied genera. The Bayesian posterior probabilities (PP > 0.9) and PhyML Bootstrap support values (BS > 50) are displayed at the nodes (PP/ML). The tree was rooted to Fusicolla violacea (CBS 634.76 T). Ex-type cultures are indicated with ‘T’, epi-type with ‘ET’, neotype with ‘NT’.
Phylogenetic analyses of different Fusarium species complexes and allied genera were conducted using different multi-locus datasets following O’Donnell et al. (2009b), Jacobs-Venter et al. (2018), Sandoval-Denis et al. (2018a, b, 2019), Lombard et al. (2019a, b), Xia et al. (2019), Crous et al. (2021), and Yilmaz et al. (2021). Briefly, phylogenetic analyses of the F. concolor, F. falsibabinda, and F. nisikadoi species complexes were performed by using the tef1-rpb1-rpb2 dataset, and rooted with F. humuli CQ1039 (Fig. 2), and single gene trees were performed respectively (Supplementary Fig. S1). Phylogenetic analyses of the F. fujikuroi species complex was performed by using the tef1-cam-rpb1-rpb2-tub2 dataset and rooted with F. nirenbergiae CBS 744.97 (Fig. 3), and single gene trees were performed respectively (Supplementary Fig. S2). A tef1-cam-rpb2 dataset was constructed for phylogenetic analyses of the F. incarnatum-equiseti species complex and rooted with F. concolor NRRL 13994 (Fig. 4). Phylogeny of the F. lateritium species complex was performed using the tef1-rpb1-rpb2-tub2 dataset, and rooted with F. sublunatum NRRL 13384 (Fig. 5). Phylogenetic analyses of the F. oxysporum species complex was performed by using the tef1-cam-rpb1-rpb2-tub2 dataset and rooted with F. globosum NRRL 26131 (Fig. 6). Phylogenetic analyses of the F. sambucinum species complex was performed using tef1-rpb1-rpb dataset and rooted with F. lactis CBS 411.97 (Fig. 7). Phylogenetic analyses of the F. tricinctum species complex was performed by using a combined ITS-tef1-rpb1-rpb2 dataset and rooted with F. concolor NRRL 13994 (Fig. 8), and single gene trees were performed respectively (Supplementary Fig. S3). Phylogenetic analyses of the genus Bisifusarium were performed by using the ITS-tef1-cam-rpb2-tub2 dataset and rooted by Rectifusarium robinianum CBS 430.91 (Fig. 9), and single gene trees were performed respectively (Supplementary Fig. S4). Phylogenetic analyses of Neocosmospora were performed using ITS-tef1-rpb2 dataset, and rooted by Geejayessia cicatricum CBS 125552 and G. atrofusca NRRL 22316 (Fig. 10), and single gene trees were performed respectively (Supplementary Fig. S5). Composition of the multi-locus datasets, outgroup taxa and character numbers and the best model of each locus were listed in Table 3.
Fig. 2.
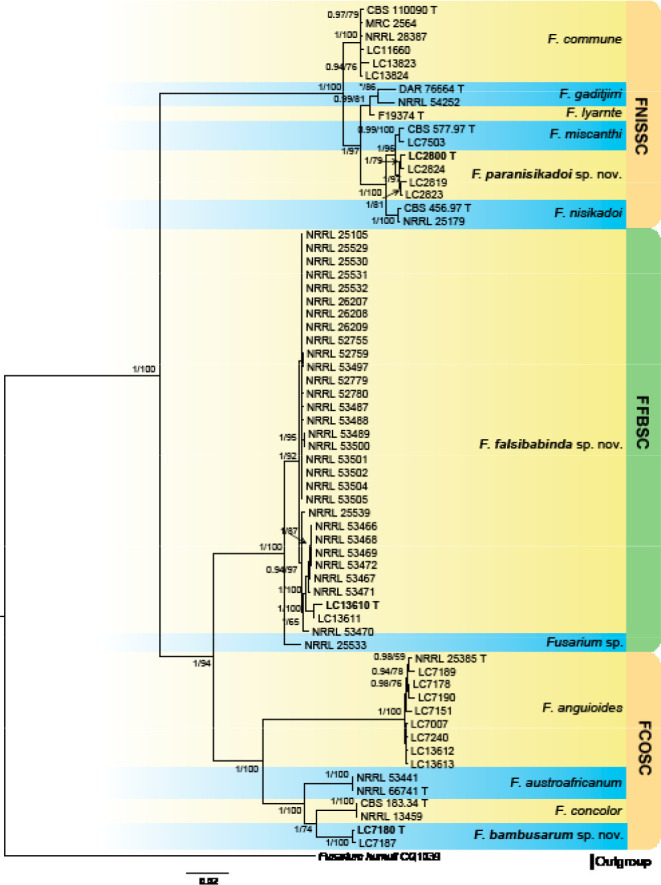
Fifty percent majority rule consensus tree from a Bayesian analysis based on a three-locus combined dataset (tef1, rpb1, and rpb2) showing the phylogenetic relationships of five species complexes within the Fusarium, namely F. concolor (FCOSC), F. falsibabinda (FFBSC), and F. nisikadoi (FNISSC). The Bayesian posterior probabilities (PP > 0.9) and PhyML Bootstrap support values (BS > 50) are displayed at the nodes (PP/ML). The tree was rooted to Fusarium humuli (CQ1039). New species are indicated in bold, ex-type cultures in bold with ‘T’.
Fig. 3.
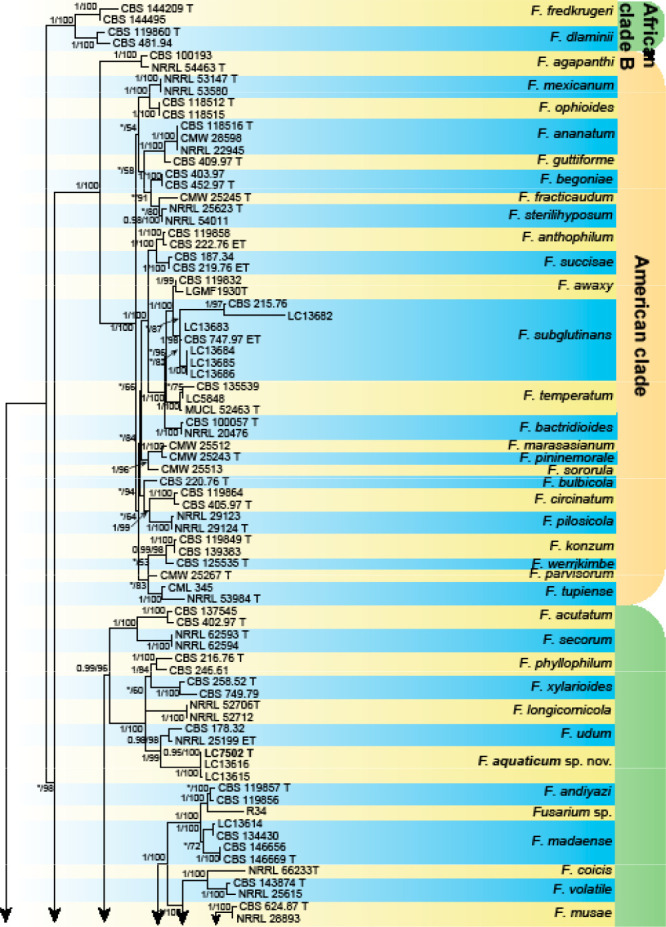
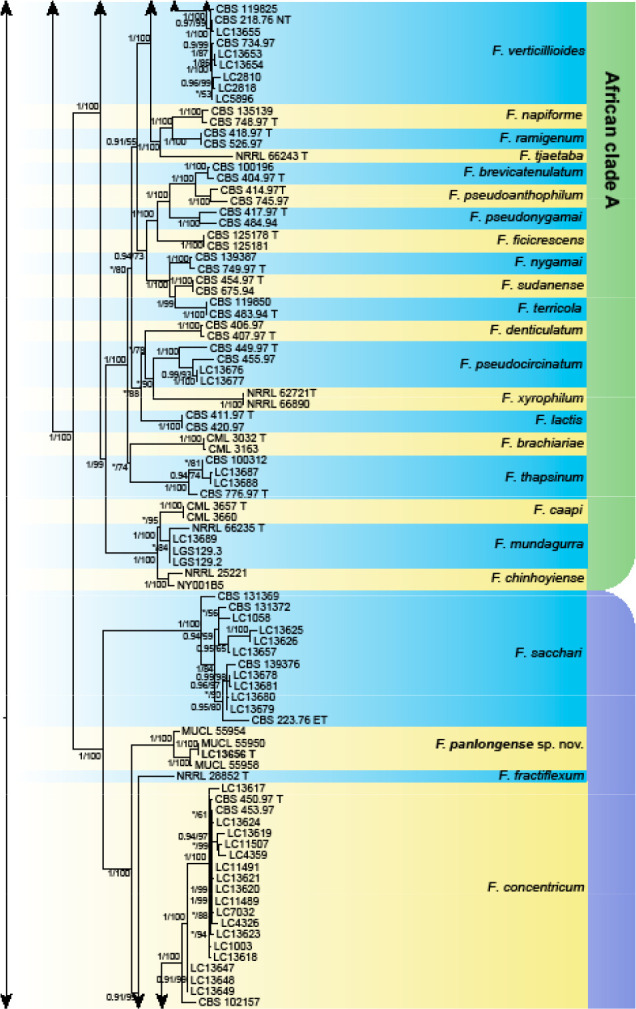
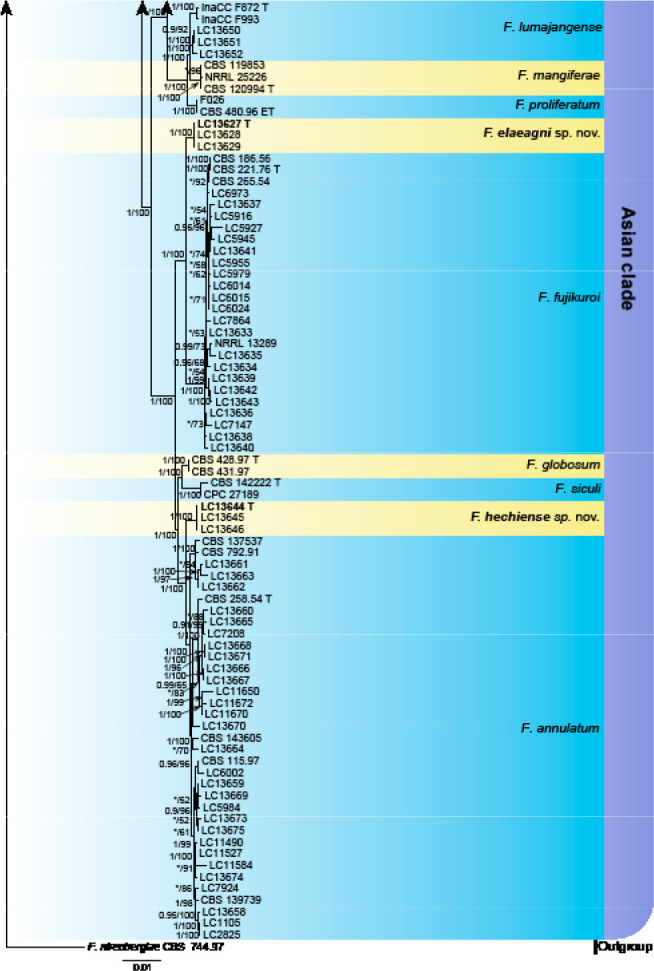
Fifty percent majority rule consensus tree from a Bayesian analysis based on a five-locus combined dataset (tef1, cam, rpb1, rpb2, and tub2) showing the phylogenetic relationships of species within the Fusarium fujikuroi species complex (FFSC). The Bayesian posterior probabilities (PP > 0.9) and PhyML Bootstrap support values (BS > 50) are displayed at the nodes (PP/ML). The tree was rooted to F. nirenbergiae (CBS 744.97). New species are indicated in bold, ex-type cultures with ‘T’, epi-type with ‘ET’, neotype with ‘NT’.
Fig. 4.
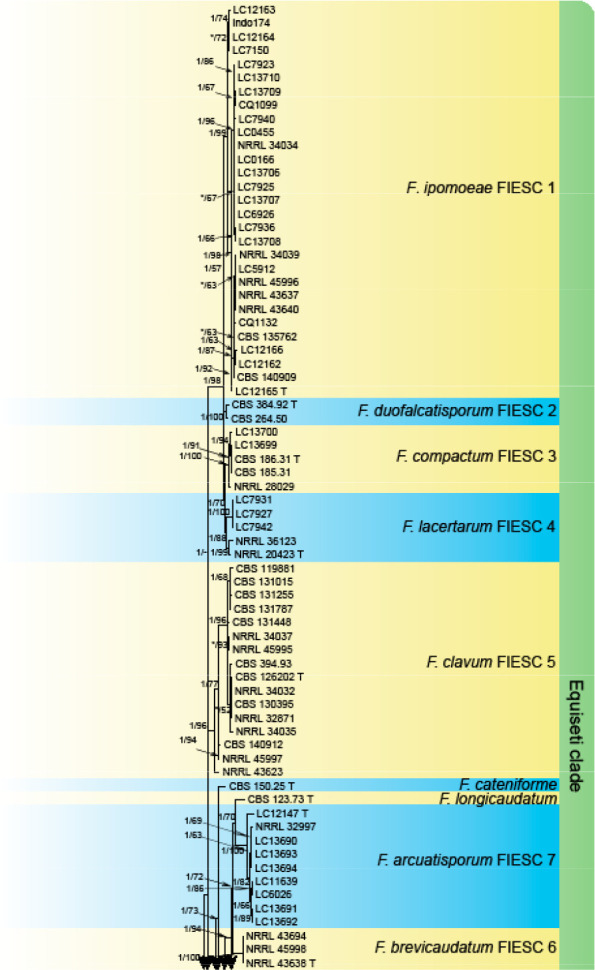
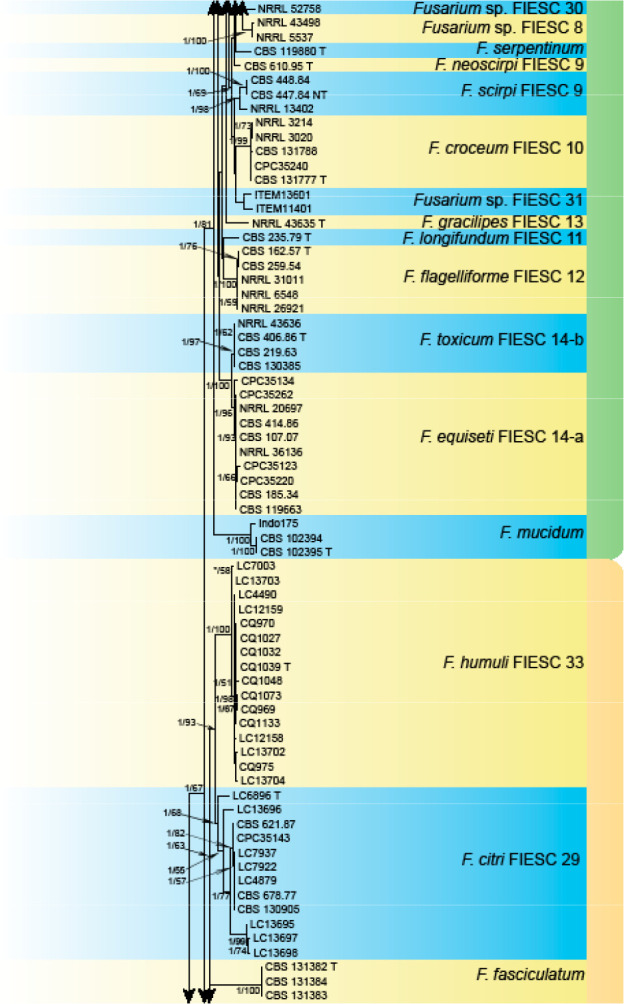
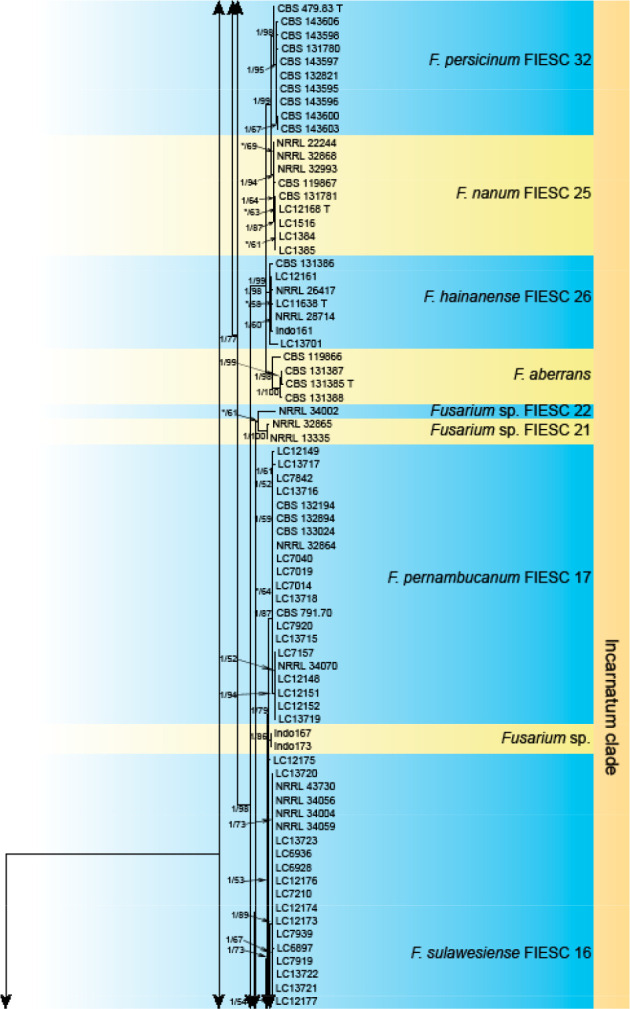
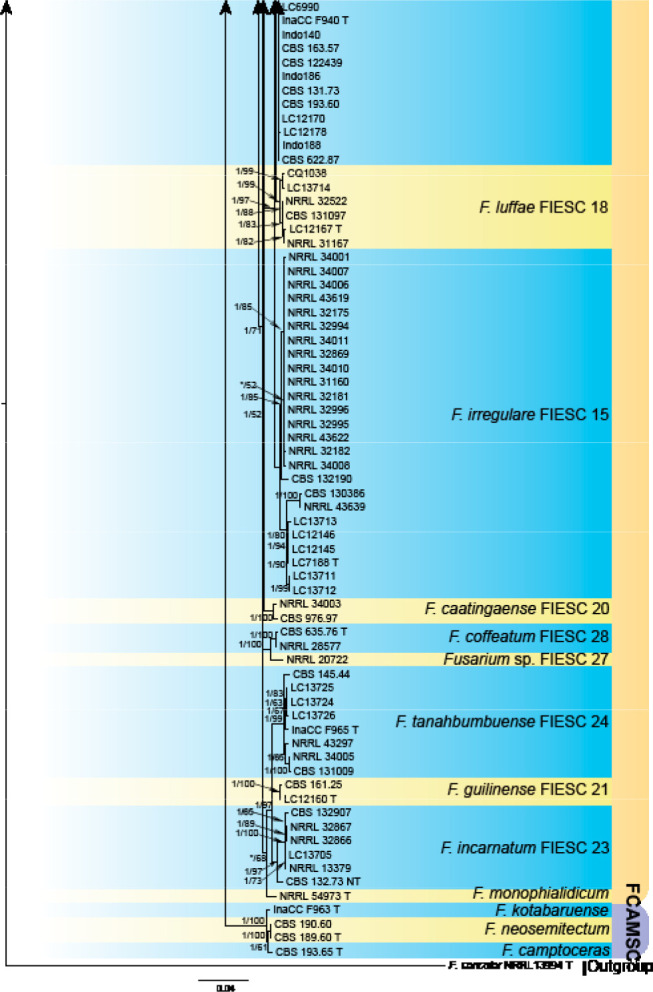
Fifty percent majority rule consensus tree from a Bayesian analysis based on a three-locus combined dataset (tef1, cam, and rpb2) showing the phylogenetic relationships of species within the Fusarium incarnatum-equiseti species complex (FIESC). The Bayesian posterior probabilities (PP > 0.9) and PhyML Bootstrap support values (BS > 50) are displayed at the nodes (PP/ML). The tree was rooted to F. concolor (NRRL 13994 T). Ex-type cultures are indicated with ‘T’, neotype with ‘NT’.
Fig. 5.
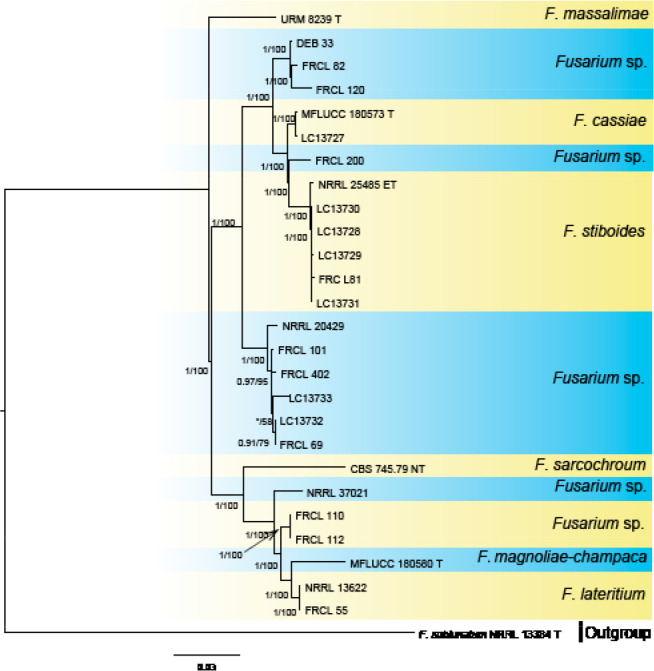
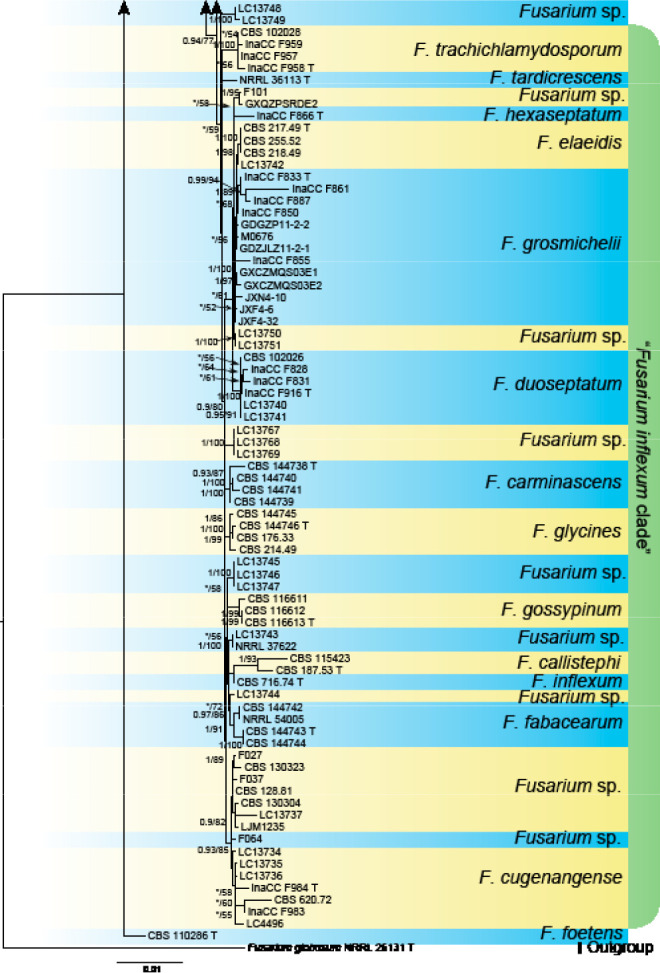
Fifty percent majority rule consensus tree from a Bayesian analysis based on a four-locus combined dataset (tef1, rpb1, rpb2, and tub2) showing the phylogenetic relationships of species within the Fusarium lateritium species complex (FLSC). The Bayesian posterior probabilities (PP > 0.9) and PhyML Bootstrap support values (BS > 50) are displayed at the nodes (PP/ML). The tree was rooted to F. sublunatum (NRRL 13384 T). Ex-type culture are indicated with ‘T’, epitype with ‘ET’, and neotype with ‘NT’.
Fig. 6.
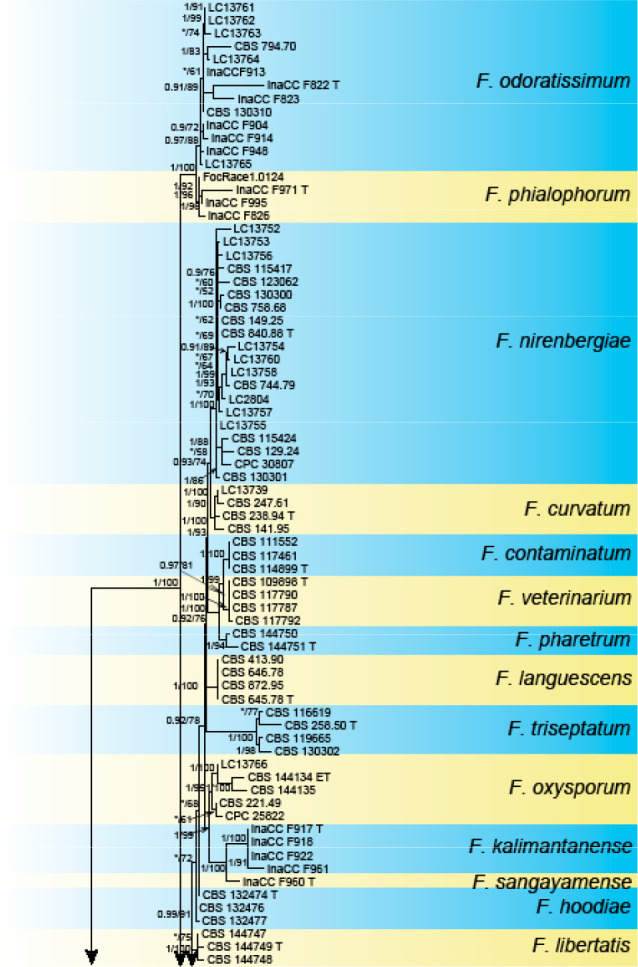
Fifty percent majority rule consensus tree from a Bayesian analysis based on a five-locus combined dataset (tef1, cam, rpb1, rpb2, and tub2) showing the phylogenetic relationships of species within the Fusarium oxysporum species complex (FOSC). The Bayesian posterior probabilities (PP > 0.9) and PhyML Bootstrap support values (BS > 50) are displayed at the nodes (PP/ML). The tree was rooted to F. globosum (NRRL 26131). Ex-type cultures are indicated with ‘T’, epitype with ‘ET’.
Fig. 7.
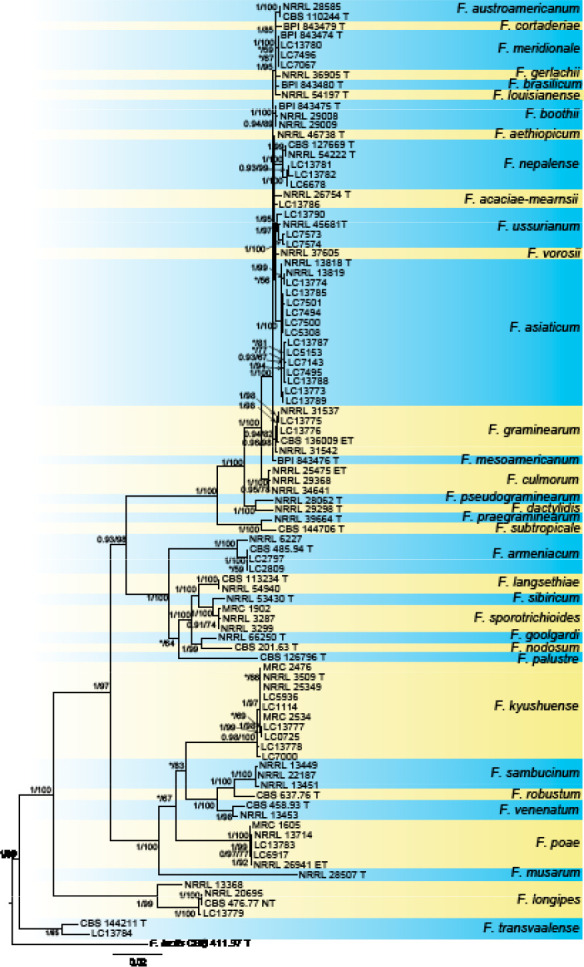
Fifty percent majority rule consensus tree from a Bayesian analysis based on a three-locus combined dataset (tef1, rpb1, and rpb2) showing the phylogenetic relationships of species within the Fusarium sambucinum species complex (FSAMSC). The Bayesian posterior probabilities (PP > 0.9) and PhyML Bootstrap support values (BS > 50) are displayed at the nodes (PP/ML). The tree was rooted to F. lactis (CBS 411.97 T). Ex-type cultures are indicated with ‘T’, epitype with ‘ET’.
Fig. 8.
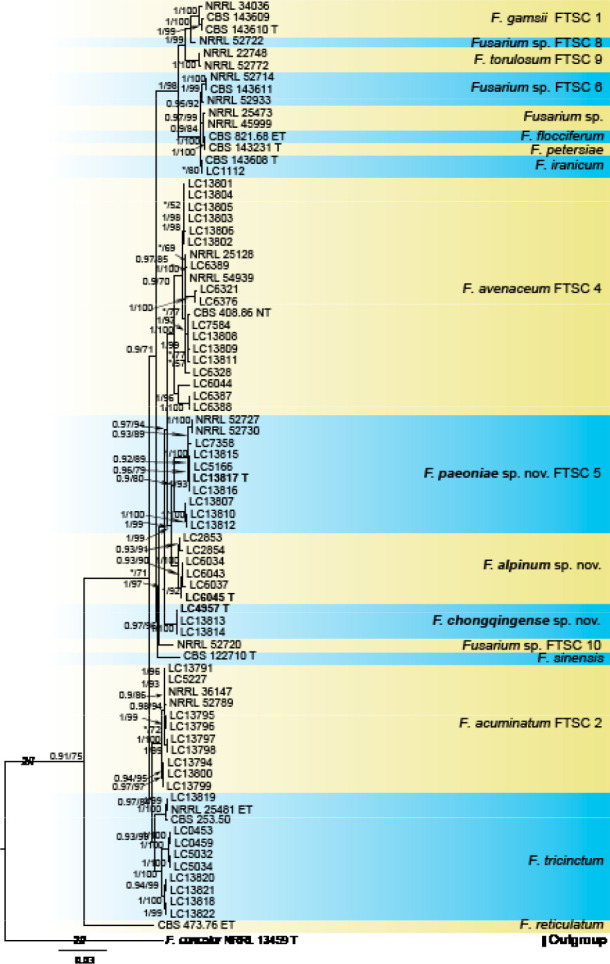
Fifty percent majority rule consensus tree from a Bayesian analysis based on a four-locus combined dataset (ITS, tef1, rpb1, and rpb2) showing the phylogenetic relationships of species within the Fusarium tricinctum species complex (FTSC). The Bayesian posterior probabilities (PP > 0.9) and PhyML Bootstrap support values (BS > 50) are displayed at the nodes (PP/ML). The tree was rooted to F. concolor (NRRL 13994 T). New species are indicated in bold, ex-type cultures are indicated with ‘T’, epitype with ‘ET’, neotype with ‘NT’.
Fig. 9.
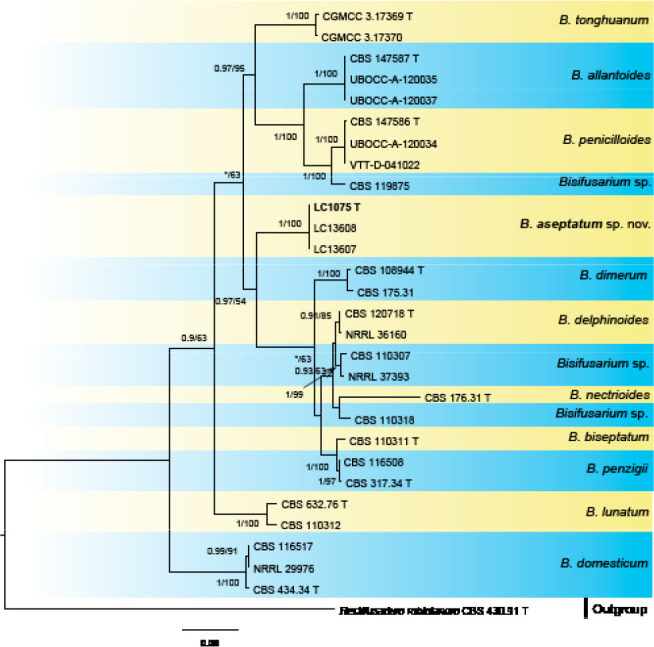
Fifty percent majority rule consensus tree from a Bayesian analysis based on a five-locus combined dataset (ITS, tef1, cam, rpb2, and tub2) showing the phylogenetic relationships of species within the Bisifusarium. The Bayesian posterior probabilities (PP > 0.9) and PhyML Bootstrap support values (BS > 50) are displayed at the nodes (PP/ML). The tree was rooted to Rectifusarium robinianum (CBS 430.91 T). New species are indicated in bold, ex-type cultures with ‘T’.
Fig. 10.
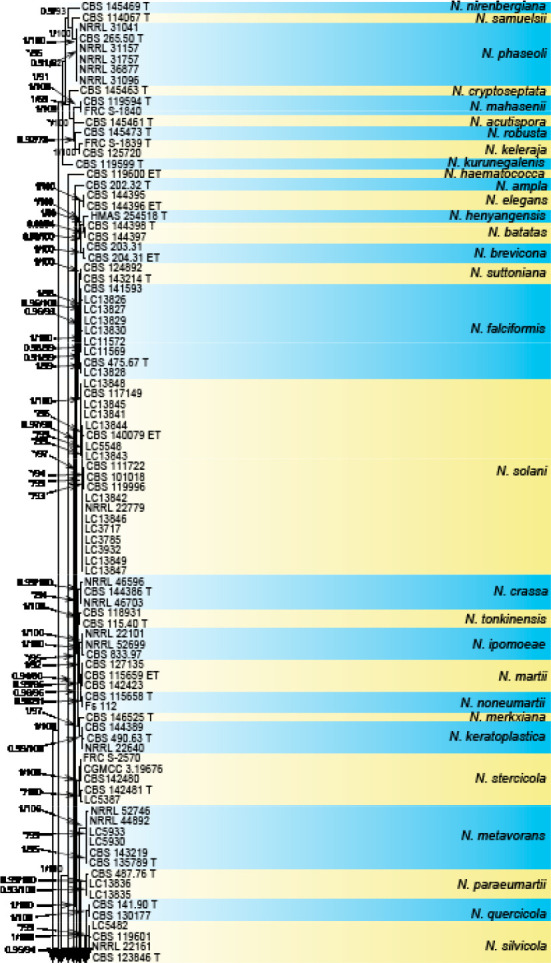
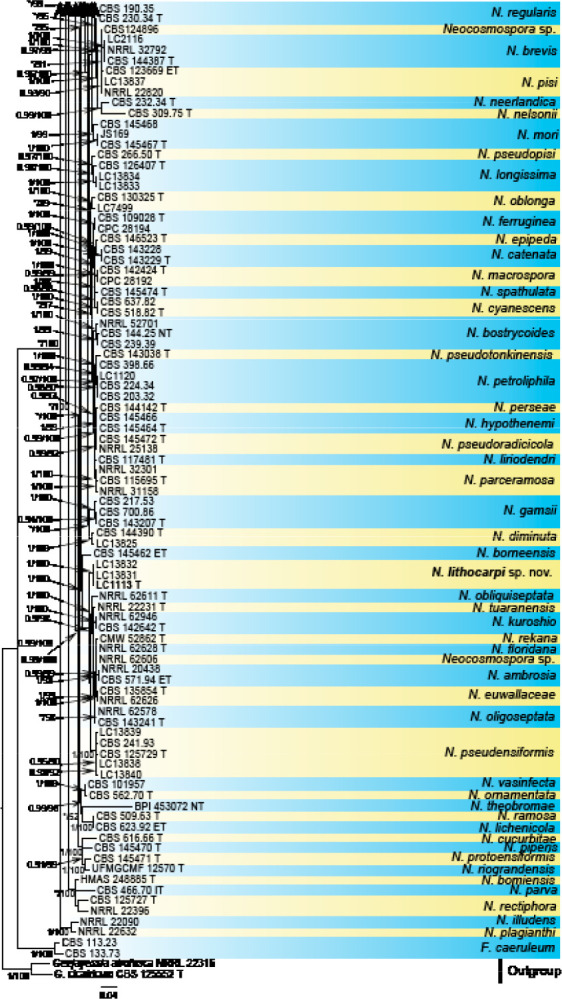
Fifty percent majority rule consensus tree from a Bayesian analysis based on a three-locus combined dataset (ITS, tef1, and rpb2) showing the phylogenetic relationships of species within the genus Neocosmospora. The Bayesian posterior probabilities (PP > 0.9) and PhyML Bootstrap support values (BS > 50) are displayed at the nodes (PP/ML). The tree was rooted to Geejayessia cicatricum (CBS 125552) and G. atrofusca (NRRL 22316). Ex-type cultures are indicated with ‘T’, epitype with ‘ET’.
Table 3.
Number of characters/model for BI analysis of each locus in phylogenetic analyses of different Fusarium species complexes and two other genera.
| Genus/Species complex | ITS | tef1 | cam | rpb1 | rpb2 | tub2 | Outgroup taxon |
|---|---|---|---|---|---|---|---|
| Bisifusarium | 480/SYM+I+G | 660/GTR+G | 565/SYM+G | – | 1455/SYM+G | 528/HKY+I+G | F. concolor |
| Fusarium concolor | – | 627/GTR+G | – | 1585/SYM+G | 1601/GTR+G | – | F. humuli |
| F. falsibabinda | – | 627/GTR+G | – | 1585/SYM+G | 1601/GTR+G | – | F. humuli |
| F. fujikuroi | – | 666/GTR+I+G | 673/SYM+I | 1549/SYM+G | 1455/SYM+I+G | 573/SYM+G | F. nirenbergiae |
| F. incarnatum-equiseti | – | 592/GTR+I+G | 547/SYM+G | – | 816/GTR+I+G | – | F. concolor |
| F. lateritium | – | 645/HKY+G | – | 1586/SYM+G | 1716/GTR+I+G | 555/HKY+G | F. sublunatum |
| F. nisikadoi | – | 627/GTR+G | – | 1585/SYM+G | 1601/GTR+G | – | F. humuli |
| F. oxysporum | – | 544/HKY+G | 552/K80 | 1455/SYM+G | 1704/GTR+G | 505/SYM+G | F. udum |
| F. sambucinum | – | 621/GTR+G | – | 1492/SYM+G | 1293/SYM+I | – | F. lactis |
| F. tricinctum | 491/SYM+I | 613/GTR+G | – | 1575/SYM+G | 1270/SYM+G | – | F. concolor |
| Neocosmospora | 333/GTR+I+G | 606/GTR+G | – | – | 1202/SYM+I+G | – | Geejayessia atrofusca and G. cicatricum |
Taxonomy
In total 425 strains were isolated. Of these, 356 isolated from China and were identified to 72 species, including 61 known and 11 novel species (Table 1). Sixty-nine isolates from diverse plants imported from 13 countries were identified as 26 species including one new species, namely F. falsibabinda. New species in Fusarium are treated alphabetically based on their respective species complexes.
FUSARIUM
Fusarium concolor species complex
Fusarium bambusarum M.M. Wang & L. Cai, sp. nov. — Myco-Bank MB 842152; Fig. 11
Fig. 11.
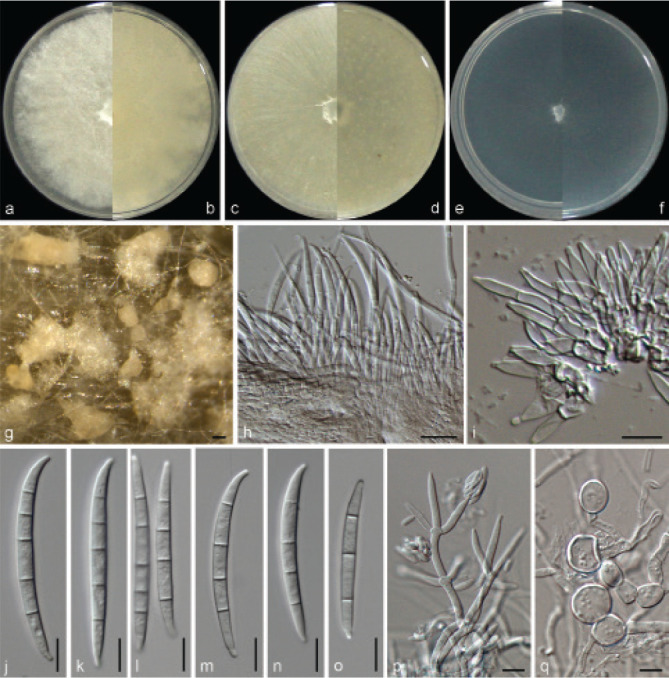
Fusarium bambusarum (ex-type culture LC7180). a, b. Colony on PDA: a. surface of colony on PDA after 7 d at 25 °C; b. reverse of colony on PDA; c–d. colony on OA: c. surface of colony on OA after 7 d at 25 °C; d. reverse of colony on OA: e–f. colony on SNA; e. surface of colony on SNA after 7 d at 25 °C; f. reverse of colony on SNA; g. sporodochia on carnation leaves; h–i. conidiophores and phialides on sporodochia; j–o. sporodochial conidia (macroconidia); p. conidiophores and phialides on aerial mycelium; q. chlamydospores. — Scale bars: g = 50 μm; h = 20 μm; i–q = 10 μm.
Etymology. Named after the host of the type specimen, bamboo.
Typus. China, Jiangxi Province, from bamboo, July 2016, J.E. Huang (HMAS 351575, holotype designated here, dried culture on SNA with carnation leaves, culture ex-type CGMCC 3.20820 = LC7180).
Colonies on PDA grown in the dark reaching 5.7–5.9 cm diam after 7 d at 25 °C, raised, aerial mycelia dense, colony margin erose to entire, surface and reverse white. Colonies on OA grown in the dark reaching 5.9–6.1 cm diam after 7 d at 25 °C, raise, aerial mycelia dense, colony margin entire, surface and reverse white. Colonies on SNA grown in the dark reaching 5.2–5.5 cm diam after 7 d at 25 °C, flat, aerial mycelia scant, colony margin erose, surface and reverse white. Pigment and odour absent. Sporodochia orange grey (5B2), formed abundantly on carnation leaves. Conidiophores in sporodochia verticillately branched and densely packed, consisting of a short, smooth- and thin-walled stipe, 4–7 × 3–5 μm, bearing an apical pair or whorls of 3 monophialides; sporodochial phialides subulate to subcylindrical, 12–15 × 3–5 μm, smooth- and thin-walled, sometimes showing a reduced and flared collarette. Sporodochial macroconidia falcate, slender, slightly curved with almost parallel sides tapering slightly towards both ends, with a papillate to hooked, curved apical cell and a foot-like basal cell, 3–6-septate, hyaline, smooth- and thin-walled; 3-septate conidia: (39.4–)41.2–50(–51.3) × 3.4–5.6 μm (av. ± sd. 45.7 ± 2.4 × 4.3 ± 0.6 μm); 4-septate conidia: (50.3–)51–59.6(–59.7) × 3.1–5.9 μm (av. ± sd. 56 ± 2.7 × 4.4 ± 0.8 μm); 5–6-septate conidia: (62.9–)63.3–85.2(–85.7) × 3.6–6.2 μm (av. ± sd. 73.2 ± 5.5 × 4.9 ± 0.7 μm). Conidiophores borne on aerial mycelia 30–110 μm tall, unbranched or sparingly branched, bearing terminal or intercalary monophialides, often reduced to single phialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, 23–30 × 3–4 μm; aerial microconidia forming small false heads on the tips of the monophialides, hyaline, oval, smooth- and thin-walled, aseptate, (5–)5.5–11(–12) × 1.6–3.5 μm (av. ± sd. 7.9 ± 1.4 × 2.8 ± 0.4 μm). Chlamydospores terminal, almost globose, rough, thick-walled, hyaline, aseptate, 6.3–12.8 μm diam (av. ± sd. 10.4 ± 2.1).
Additional material examined. China, Guangdong Province, Guangzhou city, from bamboo, July 2016, L. Cai, LC7187.
Notes — The two isolates were resolved as a strongly supported genealogically exclusive lineage in the combined tef1, rpb1, and rpb2 phylogeny (Fig. 2). Phylogenetically, F. bambusarum is closely related to F. austroafricanum and F. concolor, but differs by 152 bp and 136 bp in the three loci dataset, respectively. Morphologically, this species is distinguished based on the number of septa in sporodochial macroconidia (3–6-septate in F. bambusarum vs 0–11-septate in F. austroafricanum) and in the type of aerial phialides (monophialides in F. bambusarum vs polyphialides in F. concolor) (Marasas et al. 1986).
Fusarium falsibabinda species complex
Fusarium falsibabinda M.M. Wang & L. Cai, sp. nov. — Myco-Bank MB 842153; Fig. 12
Fig. 12.
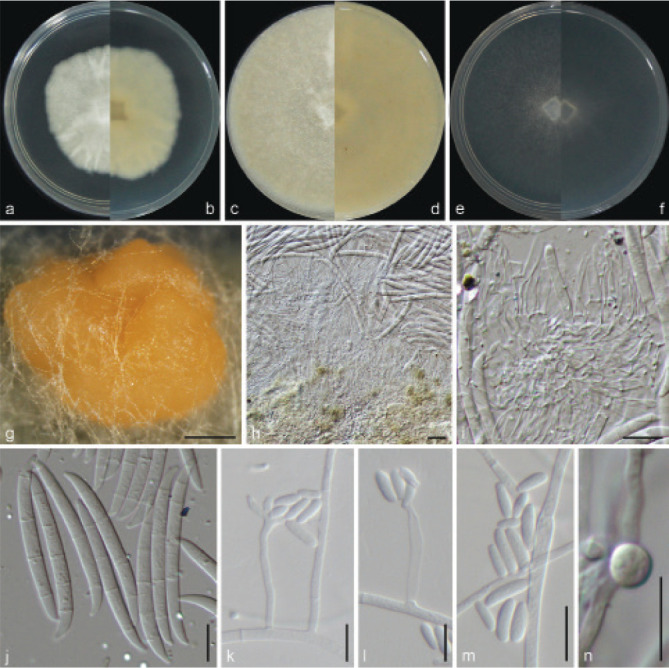
Fusarium falsibabinda (ex-type culture LC13610). a–b. Colony on PDA: a. surface of colony on PDA after 7 d at 25 °C; b. reverse of colony on PDA; c–d. colony on OA: c. surface of colony on OA after 7 d at 25 °C; d. reverse of colony on OA; e–f. colony on SNA: e. surface of colony on SNA after 7 d at 25 °C; f. reverse of colony on SNA; g. sporodochium on carnation leaves; h–i. conidiophores and phialides on sporodochia; j. sporodochial conidia (macroconidia); k–l. phialides on aerial mycelium; m. aerial conidia (microconidia); n. chlamydospores. — Scale bars: g = 50 μm; h = 20 μm; i–n = 10 μm.
Etymology. Named after species of this newly introduced clade, F. babinda.
Typus. Japan, intercepted and isolated at Ningbo Customs, from Podocarpus macrophyllus imported to China, Oct. 2012, W.J. Duan (HMAS 351576, holotype designated here, dried culture on SNA with carnation leaves, culture ex-type CGMCC 3.20823 = LC13610 = F015).
Colonies on PDA grown in the dark reaching 3.7–4.2 cm diam after 7 d at 25 °C, raised, aerial mycelia dense, colony margin erose, surface white; reverse pale yellow in the centre, white at the margin. Colonies on OA grown in the dark reaching 5.9– 6.1 cm diam after 7 d at 25 °C, raised, aerial mycelia dense, colony margin entire, surface and reverse white. Colonies on SNA grown in the dark reaching 5.2–5.5 cm diam after 7 d at 25 °C, flat, aerial mycelia scant, colony margin erose, surface and reverse white. Pigment and odour absent. Sporodochia golden yellow (5B7), formed abundantly on carnation leaves. Conidiophores in sporodochia verticillately branched and densely packed, consisting of a smooth- and thin-walled stipe, 14–17 × 5–6 μm, bearing apical pairs or whorls of 3 monophialides; sporodochial phialides subulate to subcylindrical, 15–21 × 3–5 μm, smooth- and thin-walled, sometimes showing a reduced and flared collarette. Sporodochial macroconidia falcate, slender, slightly curved with almost parallel sides tapering slightly towards both ends, with a papillate to hooked apical cell and a barely notched to distinctly notched basal cell, 3–5-septate, hyaline, smooth- and thin-walled; 3-septate conidia: (39.4–) 41.2–47(–49.3) × 3.4–4.5 μm (av. ± sd. 44.6 ± 2.4 × 3.9 ± 0.6 μm); 4-septate conidia: (42.3–)44–49.6(–51.7) × 3.6–4.5 μm (av. ± sd. 47 ± 1.7 × 4.1 ± 0.4 μm); 5-septate conidia: (50.9–) 51.3–53.2(–53.5) × 3.7–4.5 μm (av. ± sd. 52.2 ± 0.8 × 4.2 ± 0.3 μm). Conidiophores borne on aerial mycelia 30–50 μm tall, unbranched, polyphialides or monophialides, often reduced to single phialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, 30–40 × 3–5 μm; aerial microconidia forming small false heads on tips of mono- and polyphialides, hyaline, oval or obovoid with a truncate base, smooth- and thin-walled, aseptate, (6–)6.5–11(–12) × 2.6–3.5 μm (av. ± sd. 7.9 ± 1.4 × 3.1 ± 0.4 μm). Chlamydospores intercalary, almost globose, slight rough, thick-walled, hyaline, aseptate, 4.3–5.1 μm diam (av. ± sd. 4.7 ± 0.3).
Additional material examined. Japan, intercepted and isolated at Ningbo Customs, from Camellia sasanqua imported to China, Mar. 2014, W.J. Duan, LC13611 (= F058).
Notes — Several strains isolated from soil in China (NRRL 25539, NRRL 53467, and NRRL 53470), and Camellia sasanqua and Podocarpus macrophyllus from Japan (LC13610 and LC13611), clustered as a distinct clade near the F. concolor complex (Fig. 2). This clade was recognised as F. babinda by Jacobs-Venter et al. (2018) and Sandoval-Denis et al. (2018a), with NRRL 25539 (= CBS 396.96) as the representative isolate. However, based on the ex-type isolate of F. babinda (BBA 69872 = F11217 = NRRL 25807) designated in Summerell et al. (1995), Crous et al. (2021) confirmed that F. babinda clustered in the F. fujikuroi complex, distant from the clade encompassing NRRL 25539. In this paper, we introduce a new species, F. falsibabinda, to represent this previously incorrectly named clade (Fig. 2). Based on morphology, F. falsibabinda is distinct from F. babinda in the sporodochia colour (golden yellow in F. falsibabinda vs pale orange in F. babinda), macroconidial size (39.4–53.5 × 3.4–4.5 μm in F. falsibabinda vs 32–72 × 4–6 μm in F. babinda), type of conidiophores (polyphialides or monophialides in F. falsibabinda vs monophialides in F. babinda), and shape and septation of microconidia (oval or obovoid with a truncate base, aseptate in F. falsibabinda vs fusiform, 0–1-septate in F. babinda) (Summerell et al. 1995, Leslie & Summerell 2006). Phylogenetically, F. falsibabinda is closest to an undescribed Fusarium species (represented by NRRL 25533), with both taxa residing in the F. falsibabinda species complex (Fig. 2).
Fusarium fujikuroi species complex
Fusarium aquaticum M.M. Wang & L. Cai, sp. nov. — Myco-Bank MB 842154; Fig. 13
Fig. 13.
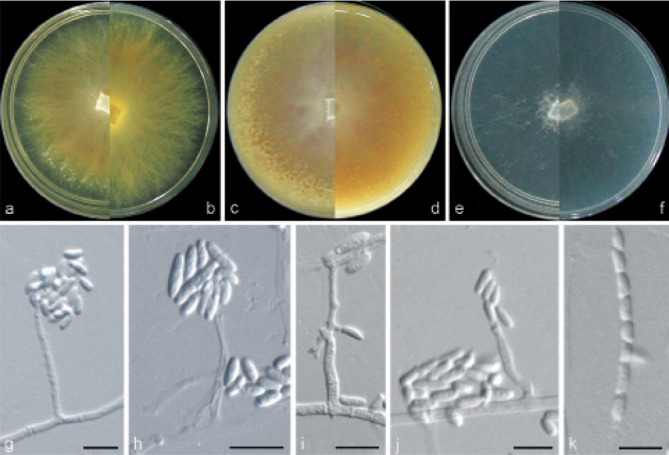
Fusarium aquaticum (ex-type culture LC7502). a–b. Colony on PDA: a. surface of colony on PDA after 7 d at 25 °C; b. reverse of colony on PDA; c–d. colony on OA: c. surface of colony on OA after 7 d at 25 °C; d. reverse of colony on OA; e–f. colony on SNA: e. surface of colony on SNA after 7 d at 25 °C; f. reverse of colony on SNA; g–j. conidiophores and phialides on aerial mycelium; k. aerial conidia. — Scale bars: g–k = 10 μm.
Etymology. Refers to its habitat, water, from which the holotype was isolated.
Typus. China, Guizhou Province, Zunyi city, from water, May 2015, L. Cai, Z.F. Zhang, X. Zhou & J.R. Jiang (HMAS 351577, holotype designated here, dried culture on SNA with carnation leaves; culture ex-type CGMCC 3.20819 = LC7502). Colonies on PDA grown in the dark reaching 5.7–5.9 cm diam after 7 d at 25 °C, flat, aerial mycelia scant, colony margin filamentous to erose, filiform, surface pastel yellow (2A4) in the centre, white at the margin; reverse pastel yellow (2A4). Colonies on OA grown in the dark reaching 5.8–6.2 cm diam after 7 d at 25 °C, flat, aerial mycelia dense, colony margin entire, surface and reverse pastel yellow (3A4). Colonies on SNA grown in the dark reaching 5.4–5.7 cm diam after 7 d at 25 °C, flat, aerial mycelia scant, colony margin erose, white; reverse white. Pigment and odour absent. Sporodochia not observed. Conidiophores borne on aerial mycelia 30–50 μm tall, unbranched or rarely branched, bearing terminal or intercalary mono- or polyphialides, often reduced to single phialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, 2–23 × 2.5–3 μm, periclinal thickening inconspicuous or absent; aerial microconidia single, forming short chains or small false heads on tips of mono- and polyphialides, hyaline, ovoid, ellipsoid to reniform, smooth- and thin-walled, aseptate, (4–)4.1–11.9(–12.7) × 1.6–3.7 μm (av. ± sd. 6.9 ± 2 × 2.6 ± 0.5 μm). Chlamydospores not observed.
Additional material examined. China, Guizhou Province, Zunyi city, from water, May 2015, L. Cai, Z.F. Zhang, X. Zhou & J.R. Jiang, LC13615; ibid., LC13616.
Notes — Fusarium aquaticum is phylogenetically closely related to F. udum (Fig. 3), but differs by 68 bp in the five loci dataset. Morphologically, F. aquaticum is distinct from F. udum in the type of aerial phialides (polyphialides or monophialides in F. aquaticum vs monophialides in F. udum), shape and septation of aerial microconidia (ovoid, ellipsoid to reniform, aseptate in F. aquaticum vs fusoid to reniform or ovoid 0–1-septate in F. udum) (Leslie & Summerell 2006).
Fusarium elaeagni M.M. Wang & L. Cai, sp. nov. — MycoBank MB 842155; Fig. 14
Fig. 14.
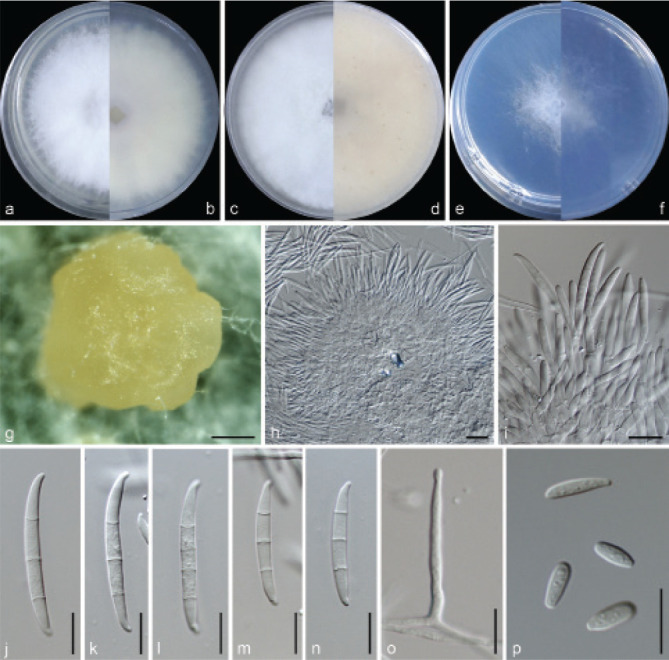
Fusarium elaeagni (ex-type culture LC13627). a–b. Colony on PDA; a. surface of colony on PDA after 7 d at 25 °C; b. reverse of colony on PDA; c–d. colony on OA: c. surface of colony on OA after 7 d at 25 °C; d. reverse of colony on OA; e–f. colony on SNA: e. surface of colony on SNA after 7 d at 25 °C; f. reverse of colony on SNA; g. sporodochium on carnation leaves; h–i. conidiophores and phialides on sporodochia; j–n. sporodochial conidia (macroconidia); o. conidiophores and phialides on aerial mycelium; p. aerial conidia. — Scale bars: g = 50 μm; h = 20 μm; i–q = 10 μm.
Etymology. Named after the host genus of the type specimen, Elaeagnus.
Typus. China, Jiangsu Province, Suzhou city, from Elaeagnus pungens, Nov. 2017, Q. Chen (HMAS 351578, holotype designated here, dried culture on SNA with carnation leaves; culture ex-type CGMCC 3.20822 = LC13627 = CQ1053).
Colonies on PDA grown in the dark reaching 5.7–5.9 cm diam after 7 d at 25 °C, raised, aerial mycelia dense, colony margin erose, surface and reverse white. Colonies on OA grown in the dark, reaching 5.7–5.9 cm diam after 7 d at 25 °C, raised, aerial mycelia dense, colony margin entire, surface and reverse white. Colonies on SNA grown in the dark reaching 5.5–5.8 cm diam after 7 d at 25 °C, flat, aerial mycelia scant, colony margin erose, surface and reverse white. Pigment and odour absent. Sporodochia greyish orange (2C3), formed abundantly on carnation leaves. Conidiophores in sporodochia verticillately branched and densely packed; sporodochial phialides subulate to subcylindrical, 13–17 × 3–4 μm, smooth- and thin-walled. Sporodochial macroconidia slender, falcate, slightly curved with almost parallel sides tapering slightly towards both ends, with a papillate to hooked, curved apical cell and a blunt to foot-like basal cell, 3–4-septate, hyaline, smooth- and thin-walled, (21–)23.5–35.8(–37) × 2.5–3.7 μm (av. ± sd. 30.7 ± 4.1 × 3.1 ± 0.9 μm). Conidiophores borne on aerial mycelia 20–40 μm tall, often reduced to single mono- or polyphialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, 20–29 × 2–3 μm; aerial microconidia forming small false heads on tips of mono- and polyphialides, hyaline, ellipsoid to falcate, rarely club-shaped, smooth- and thin-walled, 0–1-septate; aseptate conidia: (5–)6–9(–11) × 1.7–4.2 μm (av. ± sd. 6.9 ± 1.1 × 2.4 ± 0.5 μm); 1-septate conidia: (8–)9–17.5(–20) × 2.1–4.2 μm (av. ± sd. 13.5 ± 2.8 × 2.8 ± 0.5 μm). Chlamydospores not observed.
Additional material examined. China, Jiangsu Province, Suzhou city, from Elaeagnus pungens, Nov. 2017, Q. Chen, LC13628 (= CQ1053.2); ibid., LC13629 (= CQ1053.3).
Notes — This species is phylogenetically closely related to F. fujikuroi, but differs by 112 bp in the five loci dataset (Fig. 3). Morphologically, F. elaeagni is distinguished in sporodochial colour (greyish orange in F. elaeagni vs orange in F. fujikuroi), macroconidial septa (3–4-septate in F. elaeagni vs 3–5-septate in F. fujikuroi), microconidial shape (ellipsoidal to falcate, rarely club-shaped in F. elaeagni vs ovoid or club-shaped in F. fujikuroi ), and the type of aerial phialides (mono- or polyphialides in F. elaeagni vs polyphialides commonly in F. fujikuroi) (Nirenberg 1976, Leslie & Summerell 2006).
>Fusarium hechiense M.M. Wang & L. Cai, sp. nov. — Myco-Bank MB 842156; Fig. 15
Fig. 15.
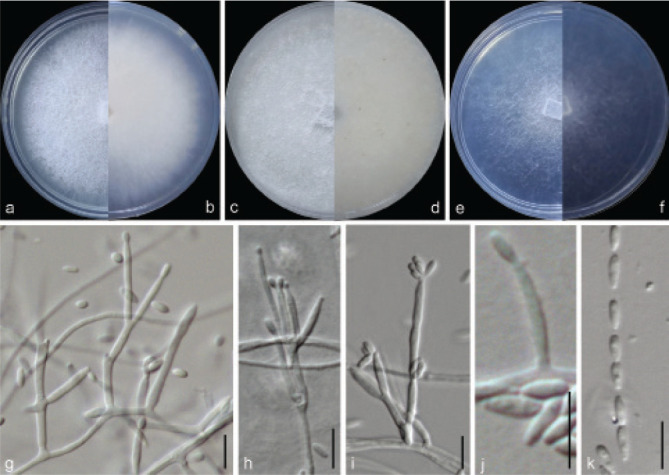
Fusarium hechiense (ex-type culture LC13644). a–b. Colony on PDA: a. surface of colony on PDA after 7 d at 25 °C; b. reverse of colony on PDA; c–d. colony on OA: c. surface of colony on OA after 7 d at 25 °C; d. reverse of colony on OA; e–f. colony on SNA: e. surface of colony on SNA after 7 d at 25 °C; f. reverse of colony on SNA; g–j. conidiophores and phialides on aerial mycelium; k. aerial conidia. — Scale bars: g–k = 10 μm.
Etymology. Named after the location of the type specimen, Hechi city.
Typus. China, Guangxi ZhuangAutonomous Region, Hechi city, Sanwang country, from Musa nana, June 2017, M.M. Wang (HMAS 351579, holotype designated here, dried culture on SNA with carnation leaves; culture ex-type CGMCC 3.20824 = LC13644 = GXHCSWL14-E1).
Colonies on PDA grown in the dark reaching 5.3–5.6 cm diam after 7 d at 25 °C, raised, aerial mycelia dense, colony margin erose, surface white; reverse yellowish white (4A2) in the centre, white at the margin. Colonies on OA grown in the dark reaching 5.7–5.9 cm diam after 7 d at 25 °C, raised, aerial mycelia dense, colony margin entire, surface and reverse white. Colonies on SNA grown in the dark reaching 5.5–5.8 cm diam after 7 d at 25 °C, flat, aerial mycelia scant, colony margin erose, white; reverse white. Pigment and odour absent. Sporodochia not observed. Conidiophores borne on aerial mycelia 15–90 μm tall, unbranched or sparingly branched, bearing terminal or intercalary monophialides, often reduced to single phialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, 15–21 × 2–4 μm, periclinal thickening inconspicuous or absent; aerial microconidia forming small false heads or chains on tips of monophialides, hyaline, subglobose, oval, reniform or obovoid with a truncate base, ellipsoidal, smooth- and thin-walled, 0–1-septate, (5–)5.2–10 × 1.8–3.5 μm (av. ± sd. 6.9 ± 1.2 × 2.6 ± 0.4 μm). Chlamydospores not observed.
Additional material examined. China, Guangxi Zhuang Autonomous Region, Hechi city, Sanwang country, from Musa nana, June 2017, M.M. Wang, LC13645 (= GXHCSWL14-E12; ibid., LC13646 (= GXHCSWL14-E13).
Notes — Fusarium hechiense is phylogenetically closely related to F. annulatum (Fig. 3), but differs by 143 bp in the five loci dataset. Morphologically, the two species are distinguished in the number of microconidial septa (0–1-septate in F. hechiense vs aseptate in F. annulatum) (Leslie & Summerell 2006).
Fusarium panlongense M.M. Wang & L. Cai, sp. nov. — Myco-Bank MB 842157; Fig. 16
Fig. 16.
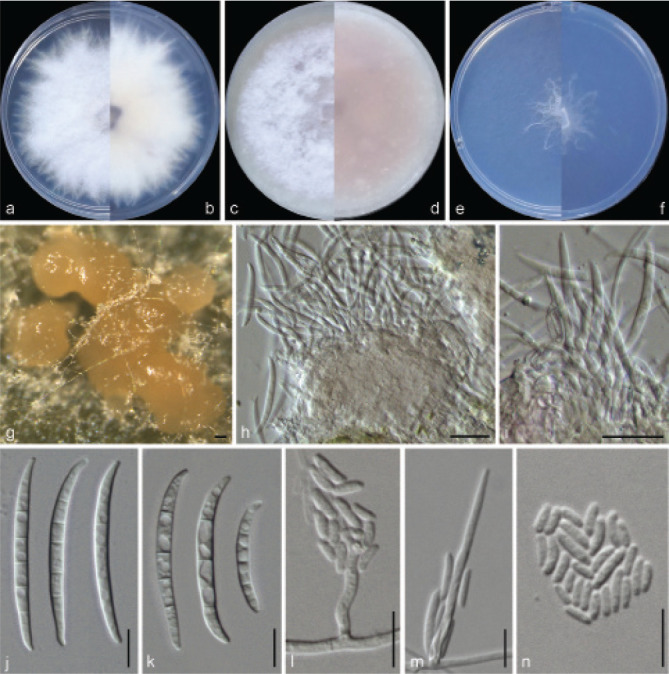
Fusarium panlongense (ex-type culture LC13656). a–b. Colony on PDA: a. surface of colony on PDA after 7 d at 25 °C; b. reverse of colony on PDA; c–d. colony on OA: c. surface of colony on OA after 7 d at 25 °C; d. reverse of colony on OA; e–f. colony on SNA: e. surface of colony on SNA after 7 d at
Etymology. Name refers to the location of the type specimen, Panlong country.
Typus. China, Guangxi Zhuang Autonomous Region, Guilin city, Panlong country, from Musa nana, June 2017, M.M. Wang (HMAS 351580, holotype designated here, dried culture on SNA with carnation leaves; culture ex-type CGMCC 3.20825 = LC13656 = GXGLPLL15E2).
Colonies on PDA grown in the dark reaching 5.7–5.9 cm diam after 7 d at 25 °C, raised, aerial mycelia dense, colony margin filamentous, erose to filiform, surface white; reverse grey (3B1) in the centre, white at the margin. Colonies on OA grown in the dark reaching 5.0–5.5 cm diam after 7 d at 25 °C, raised, aerial mycelia dense, colony margin entire, surface white; reverse orange grey (6B2) to brownish grey (6C2) in the centre, white at the margin. Colonies on SNA grown in the dark reaching 5.2–5.5 cm diam after 7 d at 25 °C, flat, aerial mycelia scant, colony margin erose, white; reverse white. Pigment and odour absent. Sporodochia brownish orange (5C4), formed abundantly on carnation leaves. Conidiophores in sporodochia verticillately branched and densely packed; sporodochial phialides subulate to subcylindrical, 9–17 × 3–4 μm, smooth- and thin-walled. Sporodochial macroconidia slender, falcate, slightly curved with almost parallel sides tapering slightly towards both ends, with a papillate to hooked, curved apical cell and a blunt to foot-like basal cell, (3–)4–5-septate, hyaline, smooth- and thin-walled; 3-septate conidia: (35–)37.4–49.7(–50.1) × 2.7–4.4 μm (av. ± sd. 41.7 ± 3.7 × 3.6 ± 0.5 μm); 4-septate conidia: (39.3–)40.3–53(–53.9) × 2.5–5.9 μm (av. ± sd. 48.4 ± 3.7 × 4 ± 0.6 μm); 5-septate conidia: (42.9–)46.1–57.5(–59.4) × 2.6–5.1 μm (av. ± sd. 51.4 ± 3.9 × 4 ± 0.6 μm). Conidiophores borne on aerial mycelia often reduced to single monophialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, 10–50 × 2–4 μm, periclinal thickening inconspicuous or absent; aerial microconidia forming small false heads on tips of monophialides, hyaline, ovoid, reniform, ellipsoid, smooth- and thin-walled, 0–1-septate; aseptate conidia: (4.3–)4.8–7.6(–8) × 25 °C; f. reverse of colony on SNA; g. sporodochia on carnation leaves; h–i. conidiophores and phialides on sporodochia; j–k. sporodochial conidia (macroconidia); l–m. phialides on aerial mycelium; n. aerial conidia (microconidia). — Scale bars: g = 50 μm; h–i = 20 μm; j–n = 10 μm. 1.5–2.7 μm (av. ± sd. 6 ± 0.7 × 2.1 ± 0.3 μm); 1-septate conidia: (7.3–)8.2–14(–16.5) × 2–3.4 μm (av. ± sd. 10.7 ± 2 × 2.7 ± 0.3 μm). Chlamydospores not observed.
Notes — Phylogenetically, F. panlongense is well separated from known species in the FFSC, and clustered basally to several species in the Asian clade of the FFSC (Fig. 3). To date all known isolates of this species were isolated from Musa spp. in China (isolates MUCL 55954, MUCL 55958, and MUCL 55950 from Hainan Province), suggesting a possible preference in host and geography. Species in the FFSC are common in Musa spp. hosts, e.g., F. concentricum, F. lumajangense, F. musae, F. sacchari, and F. verticillioides were recovered from Musa spp. from Costa Rica, Guatemala, Honduras, Indonesia, Mexico (Yilmaz et al. 2021). Fusarium panlongense was distinguished from F. concentricum in the width of macroconidia, type of aerial phialides and shape of aerial microconidia (macroconidia width 2.7–5.9 μm, monophialides, microconidia oval, reniform, ellipsoidal in F. panlongense vs macroconidia width 3.5–4 μm mono- and polyphialides, microconidia obovoid or ovoid to allantoid in F. concentricum) (Nirenberg & O’Donnell 1998), from F. lumajangense in the size of microconidia (4.3–14 × 1.5–3.4 μm in F. panlongense vs 6–23 × 2–5 μm in F. lumajangense) (Maryani et al. 2019b), from F. musae in the presence of sporodochia and macroconidia (absent in F. musae) and shape and size of aerial microconidia (ovoid, reniform, ellipsoid, 4.3–14 × 1.5–3.4 μm in F. panlongense vs claviform or ellipsoid, often truncated, 5–17 × 1.5–4 μm in F. musae) (Van Hove et al. 2011), from F. sacchari in the septation of conidia (macroconidia 3–5-septate, microconidia 0–1-septate in F. panlongense vs macroconidia usually 3-septate, microconidia 0–2-septate in F. sacchari) (Leslie & Summerell 2006), and from F. verticillioides in the shape and septation of microconidia (oval, reniform, ellipsoidal, 0–1-septate in F. panlongense vs ovoid to club-shaped with a flattened base, usually aseptate in F. sacchari) (Leslie & Summerell 2006).
Fusarium nisikadoi species complex
Fusarium paranisikadoi M.M. Wang & L. Cai, sp. nov. — MycoBank MB 842158; Fig. 17
Fig. 17.
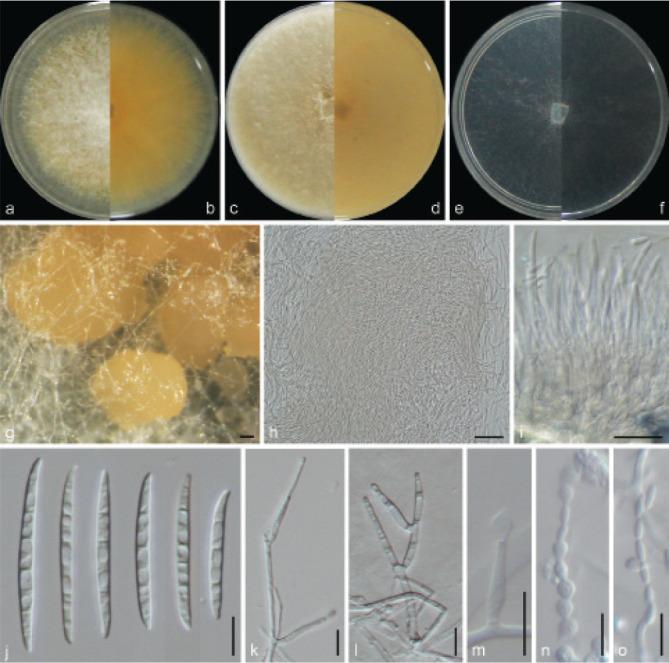
Fusarium paranisikadoi (ex-type culture LC2800). a–b. Colony on PDA: a. surface of colony on PDA after 7 d at 25 °C; b. reverse of colony on PDA; c–d. colony on OA: c. surface of colony on OA after 7 d at 25 °C; d. reverse of colony on OA; e–f. colony on SNA: e. surface of colony on SNA after 7 d at 25 °C; f. reverse of colony on SNA; g–h. sporodochia on carnation leaves; i. conidiophores and phialides on sporodochia; j. aerial conidia (macroconidia); k–m. phialides on aerial mycelium (microconidia); n. aerial conidia (napiform microconidia); o. aerial conidia (ovoid microconidia). — Scale bars: g–h = 50 μm; i = 20 μm; j–o = 10 μm.
Etymology. Named after its morphological similarity to Fusarium nisikadoi.
Typus. China, Beijing, Beijing Botanical Garden, from unidentified grass, July 2010, W. Sun (HMAS 351581, holotype designated here, dried culture on SNA with carnation leaves; culture ex-type CGMCC 3.20826 = LC2800).
Colonies on PDA grown in the dark reaching 5.7–5.9 cm diam after 7 d at 25 °C, raised, aerial mycelia dense, colony margin erose, surface greyish orange (5B3) in the centre, white at the margin; reverse greyish orange (5B4) in the centre, white at the margin. Colonies on OA grown in the dark reaching 5.9–6.2 cm diam after 7 d at 25 °C, flat, aerial mycelia scant, colony margin entire, surface orange grey (5B2) in the centre, white at the margin; reverse greyish orange (5B4) in the centre, white at the margin. Colonies on SNA grown in the dark reaching 5.7–5.9 cm diam after 7 d at 25 °C, flat, aerial mycelia scant, colony margin erose, white; reverse white. Pigment and odour absent. Sporodochia greyish orange (5B3), formed abundantly on carnation leaves. Conidiophores in sporodochia verticillately branched and densely packed, consisting of a short, smooth- and thin-walled stipe, 11–17 × 2–5 μm, bearing an apical pair or whorls of three monophialides; sporodochial phialides subulate to subcylindrical, 9.2–14.6 × 2.4–3.8 μm, smooth- and thin-walled, sometimes showing a reduced and flared collarette. Sporodochial macroconidia falcate, slightly curved with almost parallel sides tapering slightly towards both ends, with a blunt to papillate, slightly curved apical cell and a blunt to distinctly notched basal cell, 3–4-septate, hyaline, smooth- and thin-walled; 3-septate conidia: (36.7–)39.4–50.3(–51.6) × 2.3–4.1 μm (av. ± sd. 45.7 ± 3.5 × 3.1 ± 0.4 μm); 4-septate conidia: (42.8–)43.1–56.3(–57.6) × 2.5–5.2 μm (av. ± sd. 50.2 ± 3.5 × 3.8 ± 0.6 μm). Conidiophores borne on aerial mycelia, 15–80 μm tall, unbranched or sparingly branched, bearing terminal or intercalary monophialides, often reduced to single phialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, 15–25 × 2–4 μm, periclinal thickening inconspicuous or absent; aerial microconidia forming chains on the tips of the monophialides, hyaline, oval, pyriform to napiform, smooth- and thin-walled, aseptate; ovoid conidia: (3.5–)4.5–7.2(–7.7) × 1.5–3.2 μm (av. ± sd. 5.8 ± 0.9 × 2.2 ± 0.4 μm); pyriform to napiform conidia: (4.7–)5.2–8(–8.1) × 3.4–6.3 μm (av. ± sd. 6.4 ± 0.8 × 4.5 ± 0.6 μm). Chlamydospores not observed.
Additional material examined. China, Beijing, Beijing Botanical Garden, from unidentified grass, July 2010, Dimuthu, LC2819; ibid., LC2824; Beijing, Beijing Botanical Garden, from Pennisetum alopecuroides, July 2010, W. Sun, LC2823.
Notes — Fusarium paranisikadoi is phylogenetically closest to F. miscanthi and F. nisikadoi (Fig. 2), but differs from the latter by 45 bp and 71 bp in the combined tef1, rpb1, and rpb2 data-set, respectively. Morphologically, F. paranisikadoi differs from F. miscanthi in shape, septation, and size of their sporodochial macroconidia (slender, with a slightly foot-shaped basal cell and a curved and gradually tapering apical cell, 3–5-septate, 40–65(–75) × 2.5–4.5 μm in F. miscanthi vs falcate, slightly curved with almost parallel sides tapering slightly towards both ends, with a blunt to papillate, slightly curved apical cell and a blunt to distinctly notched basal cell, 3–4-septate, 36.7–57.6 × 2.3–5.2 μm in F. paranisikadoi) (Gams et al. 1999), and from F. nisikadoi in the size of their sporodochial macroconidia (56–92 × 3.5–4 μm in F. nisikadoi vs 36.7–57.6 × 2.3–5.2 μm in F. paranisikadoi) (Nirenberg & Aoki 1997).
Fusarium tricinctum species complex
Fusarium alpinum M.M. Wang & L. Cai, sp. nov. — MycoBank MB 842159; Fig. 18
Fig. 18.
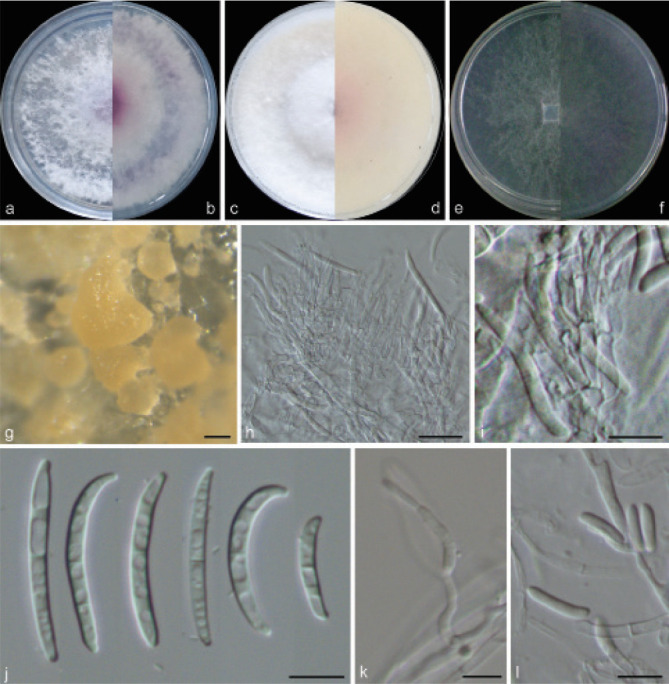
Fusarium alpinum (ex-type culture LC6045). a–b. Colony on PDA: a. surface of colony on PDA after 7 d at 25 °C; b. reverse of colony on PDA; c–d. colony on OA: c. surface of colony on OA after 7 d at 25 °C; d. reverse of colony on OA; e–f. colony on SNA: e. surface of colony on SNA after 7 d at 25 °C; f. reverse of colony on SNA; g. sporodochia on carnation leaves; h–i. conidiophores and phialides on sporodochia; j. sporodochial conidia (macroconidia); k. phialides on aerial mycelium; l. aerial conidia (microconidia). — Scale bars: g = 50 μm; h = 20 μm; i–l = 10 μm.
Etymology. Named after the special geographical reference of this species, ‘alp’.
Typus. China, TibetAutonomous Region, from species of Fabaceae, June 2015, L. Cai (HMAS 351582, holotype designated here, dried culture on SNA with carnation leaves; culture ex-type CGMCC 3.20818 = LC6045).
Colonies on PDA grown in the dark reaching 5.9–6.2 cm diam after 7 d at 25 °C, raised, punctiform, aerial mycelia dense, colony margin undulate, surface purplish grey (14C2) in the centre, white at the margin; reverse reddish lilac (14C5) in the centre, white at the margin. Colonies on OA grown in the dark reaching 5.7–5.9 cm diam after 7 d at 25 °C, raised, aerial mycelia dense, colony margin entire, surface white; reverse dull red (9B3) in the centre, white at the margin. Colonies on SNA grown in the dark reaching 5.2–5.5 cm diam after 7 d at 25 °C, flat, aerial mycelia scant, colony margin erose, white; reverse white. Pigment and odour absent. Sporodochia greyish yellow (4B4), formed abundantly on carnation leaves. Conidiophores in sporodochia verticillately branched and densely packed, consisting of a short, smooth- and thin-walled stipe, 9–11 × 3–4 μm, bearing apical whorls of 3 or more monophialides or rarely as single lateral monophialides; sporodochial phialides subulate to subcylindrical, 9.3–19.7 × 2–4 μm (av. ± sd. 13.9 ± 2.2 × 3.3 ± 0.4 μm), smooth- and thin-walled, sometimes showing a reduced and flared collarette. Sporodochial macroconidia falcate, curved slightly to dorsiventrally with almost parallel sides tapering slightly towards both ends, with a blunt to hooked, curved apical cell and a blunt to distinctly notched basal cell, 1- or 3-septate, hyaline, smooth- and thin-walled; 1-septate conidia: (15.6–)15.7–34.9(–35) × 2.4–4.6 μm (av. ± sd. 26.1 ± 6.3 × 3.3 ± 0.5 μm); 3-septate conidia: (29.2–) 30.5–46.3(–48.2) × 2.7–5.1 μm (av. ± sd. 37.8 ± 4.8 × 3.7 ± 0.6 μm). Conidiophores borne on aerial mycelia 20–70 μm tall, unbranched or sparingly branched, bearing terminal or intercalary monophialides, often reduced to single phialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, 16–23 × 2–3 μm, periclinal thickening inconspicuous or absent; aerial microconidia forming single on the tips of the monophialides, hyaline, ellipsoidal to falcate, smooth- and thin-walled, 0–1-septate; aseptate conidia: (6.8–)7.8–12.6(–12.8) × 2.2–4.7 μm (av. ± sd. 10.3 ± 1.2 × 3.9 ± 0.4 μm); 1-septate conidia: (12.7–)13.3–19.1(–20.8) × 3.1–5.5 μm (av. ± sd. 16.6 ± 1.8 × 4.4 ± 0.5 μm). Chlamydospores not observed.
Additional material examined. China, Yunnan Province, from unidentified plant, Sept. 2011, F. Liu, LC2853; ibid., LC2854; Tibet Autonomous Region, from species of Fabaceae, June 2015, L. Cai, LC6034; ibid., LC6037; ibid., LC6043.
Notes — Fusarium alpinum was collected from high altitude areas of Yunnan province and the Tibet Autonomous Region in this study. Phylogenetically, F. alpinum is closely related to F. paeoniae (Fig. 8), but differs by 44 bp in the three loci dataset. Morphologically, the two species are distinguished in the number of conidial septa (0–1(–3)-septate microconidia, 3–5-septate macroconidia in F. paeoniae vs 0–1-septate microconidia, 1- or 3-septate macroconidia in F. alpinum).
Fusarium chongqingense M.M. Wang & L. Cai, sp. nov. — MycoBank MB 842160; Fig. 19
Fig. 19.
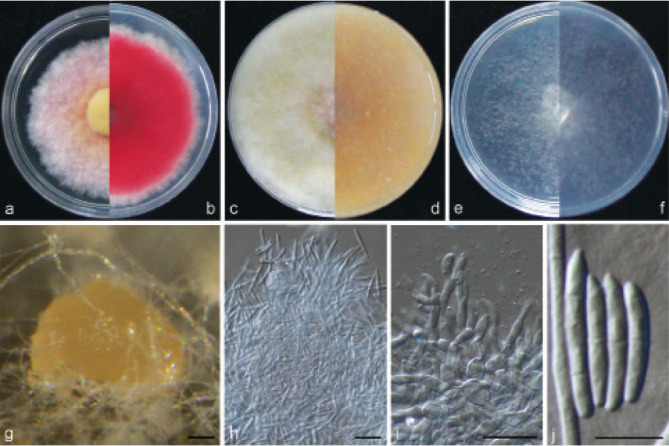
Fusarium chongqingense (ex-type culture LC4957). a–b. Colony on PDA: a. surface of colony on PDA after 7 d at 25 °C; b. reverse of colony on PDA; c–d. colony on OA: c. surface of colony on OA after 7 d at 25 °C; d. reverse of colony on OA; e–f. colony on SNA: e. surface of colony on SNA after 7 d at 25 °C; f. reverse of colony on SNA; g. sporodochium on carnation leaves; h–i. conidiophores and phialides on sporodochia; j. sporodochial conidia (macroconidia). — Scale bars: g = 50 μm; h–i = 20 μm; j = 10 μm.
Etymology. Named after the location of the type specimen, Chongqing.
Typus. China, Chongqing, Jinfo Mountain, from Bothrocaryum controversum, Oct. 2012, L. Cai (HMAS 351583, holotype designated here, dried culture on SNA with carnation leaves; culture ex-type CGMCC 3.20821 = LC4957).
Colonies on PDA grown in the dark reaching 4.6–5.1 cm diam after 7 d at 25 °C, umbonate, aerial mycelia dense, colony margin erose, surface pale yellow (4A3) to dull red (8B3) in the centre, white at the margin; reverse brownish red (10C6) in the centre, white at the margin. Colonies on OA grown in the dark reaching 5.7–5.9 cm diam after 7 d at 25 °C, flat, aerial mycelia dense, colony margin entire, surface white to greyish yellow (4C3) in the centre, white at the margin; reverse brownish orange (5C4) in the centre, white at the margin. Colonies on SNA grown in the dark reaching 5.2–5.5 cm diam after 7 d at 25 °C, flat, aerial mycelia scant, colony margin erose, white; reverse white. Pigment and odour absent. Sporodochia greyish orange (5B3), formed on carnation leaves. Conidiophores in sporodochia verticillately branched and densely packed, bearing apical pairs or whorls of three monophialides or single terminal monophialides; sporodochial phialides subulate to subcylindrical, 8–11 × 2–4 μm, smooth- and thin-walled, sometimes showing a reduced and flared collarette. Sporodochial macroconidia falcate, curved slightly with almost parallel sides tapering slightly towards both ends, with a blunt apical cell and a blunt basal cell, 1- or 3-septate, hyaline, smooth- and thin-walled; 1-septate conidia: (5.8–)8.7–18.8(–19) × 1.5–4.4 μm (av. ± sd. 13.9 ± 3 × 3.1 ± 0.4 μm); 3-septate conidia: (21–) 21.8–31.6(–31.8) × 2.6–5 μm (av. ± sd. 25.7 ± 2.5 × 4 ± 0.4 μm). Conidiophores borne on aerial mycelia not observed. Chlamydospores not observed.
Additional material examined. China, Chongqing, Jinfo Mountain, from Bothrocaryum controversum, Oct. 2012, L. Cai, LC13813; ibid., LC13814.
Notes — Fusarium chongqingense is phylogenetically closely related to F. avenaceum, F. paeoniae, and F. alpinum (Fig. 8). However, F. chongqingense differs by 67 bp from F. paeoniae, and 59 bp from F. alpinum in the three loci dataset, respectively. Morphologically, F. chongqingense is distinct based on the type of apical and basal cells of its macroconidia (blunt apical and basal cell in F. chongqingense vs long and tapering to a point to somewhat bent apical cell, and poorly to well-developed foot-shaped basal cell in F. avenaceum, blunt to papillate, curved apical cell and a blunt to foot-like basal cell in F. paeoniae; and blunt to hooked, curved apical cell and a blunt to distinctly notched basal cell in F. alpinum) (Wollenweber & Reinking 1935, Leslie & Summerell 2006).
Fusarium paeoniae M.M. Wang & L. Cai, sp. nov. — MycoBank MB 842161; Fig. 20
Fig. 20.
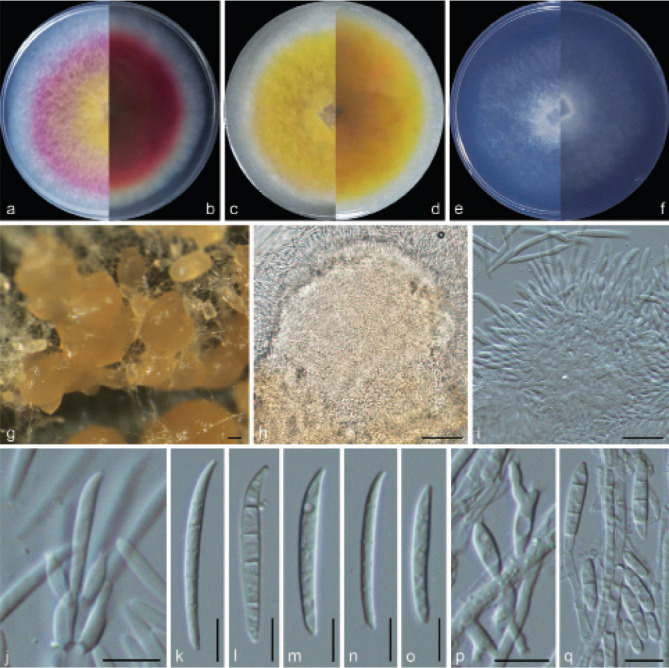
Fusarium paeoniae (ex-type culture LC13817). a–b. Colony on PDA: a. surface of colony on PDA after 7 d at 25 °C; b. reverse of colony on PDA; c–d. colony on OA: c. surface of colony on OA after 7 d at 25 °C; d. reverse of colony on OA; e–f. colony on SNA: e. surface of colony on SNA after 7 d at 25 °C; f. reverse of colony on SNA; g–h. sporodochia on carnation leaves; i–j. conidiophores and phialides on sporodochia; k–o. sporodochial conidia (macroconidia); p. phialides on aerial mycelium; q. aerial conidia (microconidia). — Scale bars: g–h = 50 μm; i–j = 20 μm; k–q = 10 μm.
Etymology. Named after the host genus of the type specimen, Paeonia.
Typus. China, Qinghai Province, from Paeonia lactiflora, Aug. 2019, M.M. Wang (HMAS 351584, holotype designated here, dried culture on SNA with carnation leaves; culture ex-type CGMCC 3.20817 = LC13817 = YZG12-2).
Colonies on PDA grown in the dark reaching 5.3–5.5 cm diam after 7 d at 25 °C, raised, aerial mycelia dense, colony margin entire, surface greyish yellow (3B4) to bluish red (12A3) in the centre, white at the margin; reverse greyish ruby (12E4) in the centre, white at the margin. Colonies on OA grown in the dark reaching 5–5.3 cm diam after 7 d at 25 °C, raised, aerial mycelia dense, colony margin entire, surface greyish yellow (3B4) in the centre, white at the margin; reverse golden brown (5D7) to greyish yellow (3B4) in the centre, white at the margin. Colonies on SNA grown in the dark reaching 4.8–5.3 cm diam after 7 d at 25 °C, flat, aerial mycelia scant, colony margin erose, white; reverse white. Pigment and odour absent. Sporodochia pale orange (5A3) to brownish orange (5C4), formed abundantly on carnation leaves. Conidiophores in sporodochia verticillately branched and densely packed, consisting of a short, smooth- and thin-walled stipe, 8–10 × 6–8 μm, bearing apical pairs or whorls of three monophialides, or as single lateral monophialides; sporodochial phialides subulate to subcylindrical, 6.9–13.3 × 2.2–4.5 μm (av. ± sd. 9.6 ± 1.7 × 3.6 ± 0.5 μm), smooth- and thin-walled, sometimes showing a reduced and flared collarette. Sporodochial macroconidia falcate, slightly curved, with a blunt to papillate, curved apical cell and a blunt to foot-like basal cell, 3–5-septate, hyaline, smooth- and thin- walled; 3-septate conidia: (27.6–)28.4–39(–39.4) × 3.8–5.8 μm (av. ± sd. 32.1 ± 3.3 × 4.5 ± 0.5 μm); 4-septate conidia: (30.3–) 32.1–41.7(–43) × 3.6–7.1 μm (av. ± sd. 37.9 ± 2.7 × 5 ± 0.8 μm); 5-septate conidia: (39.5–)39.8–50.2(–52.2) × 3.2–5.8 μm (av. ± sd. 45.2 ± 2.8 × 4.7 ± 0.7 μm). Conidiophores borne on aerial mycelia often reduced to single phialides, mono- or polyphialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, 5–20 × 3–5 μm, periclinal thickening inconspicuous or absent; aerial microconidia forming small false heads on the tips of the mono- and polyphialides, hyaline, ellipsoid to falcate, smooth- and thin-walled, 0–1(–3)-septate; aseptate conidia: (6–)7–10(–11) × 2.2–3.6 μm (av. ± sd. 8.6 ± 0.9 × 2.9 ± 0.4 μm); 1-septate conidia: (12.7–)13–15.8(–16.2) × 3.5–4.7 μm (av. ± sd. 14.2 ± 0.8 × 4.1 ± 0.3 μm); 3-septate conidia: (20.2–)21.3–25.2(–25.4) × 3.6–5.7 μm (av. ± sd. 23.4 ± 1.5 × 4.8 ± 0.6 μm). Chlamydospores not observed.
Additional material examined. China, Qinghai Province, from Crataegus monogyna, Sept. 2013, Q. Chen, LC5166; Qinghai Province, from Elymus dahuricus, Aug. 2019, M. Gao, LC13815 (= GM56); ibid., from Plantago sp., LC13807 (= GM123); ibid., from Gentiana scabra, LC13810 (= GM65); ibid., LC13812 (= GM85); Qinghai Province, from Populus sp., Aug. 2019. M.M. Wang, LC13816 (= YZG10-2); Tibet Autonomous Region, from species of Poaceae, June 2015, F. Liu, LC7358.
Notes — Phylogenetically F. paeoniae is closely related to F. alpinum (Fig. 8), but differs by 44 bp in the three loci dataset. Morphologically, the two species differ in the number of conidial septa (0–1(–3)-septate microconidia, 3–5-septate macroconidia in F. paeoniae vs 0–1-septate microconidia, 1- or 3-septate macroconidia in F. alpinum).
Bisifusarium L. Lombard et al., Stud. Mycol. 80: 223. 2015
Bisifusarium aseptatum M.M. Wang & L. Cai, sp. nov. — Myco-Bank MB 842162; Fig. 21
Fig. 21.
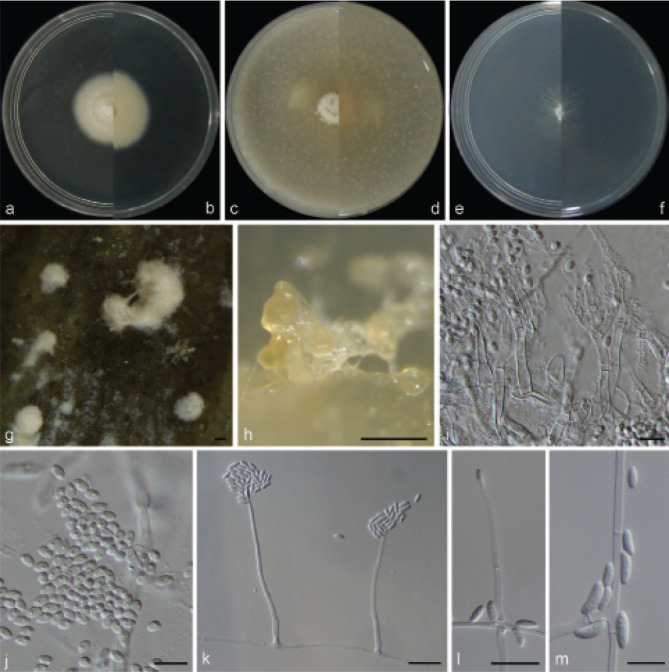
Bisifusarium aseptatum (ex-type culture LC1075). a–b. Colony on PDA: a. surface of colony on PDA after 7 d at 25 °C; b. reverse of colony on PDA; c–d. colony on OA: c. surface of colony on OA after 7 d at 25 °C; d. reverse of colony on OA; e–f. colony on SNA: e. surface of colony on SNA after 7 d at 25 °C; f. reverse of colony on SNA; g–h. sporodochia on carnation leaves; i. conidiophores and phialides on sporodochia; j. sporodochial conidia (macroconidia); k–l. phialides on aerial mycelium; m. aerial conidia (microconidia). — Scale bars: g–h = 50 μm; i–j, m = 10 μm; k–l = 20 μm.
Etymology. Refers to the aseptate sporodochial conidia.
Typus. China, Guangdong Province, Guangzhou city, from species of Orchidaceae, Mar. 2011, Y.Y. Su (HMAS 351585, holotype designated here, dried culture on SNA with carnation leaves; culture ex-type CGMCC 3.20816 = LC1075).
Colonies on PDA grown in the dark reaching 1.7–2.1 cm diam after 7 d at 25 °C, flat, aerial mycelia dense, colony margin erose, surface and reverse white. Colonies on OA grown in the dark reaching 0.9–1.1 cm diam after 7 d at 25 °C, flat, aerial mycelia dense, colony margin entire, surface and reverse white. Colonies on SNA grown in the dark reaching 1.2–1.5 cm diam after 7 d at 25 °C, flat, aerial mycelia scant, colony margin filamentous, white; reverse white. Pigment and odour absent. Sporodochia white to yellowish white (4A3), formed on carnation leaves. Conidiophores in sporodochia forming a smooth- and thin-walled stipe, bearing apical whorls of mostly 3 monophialides; sporodochial phialides subulate to subcylindrical, 8–10 × 3–4 μm, smooth- and thin-walled. Sporodochial macroconidia oval, reniform, aseptate, hyaline, smooth- and thin-walled; (4.4–) 4.5–7(–7.1) × 2.6–4.1 μm (av. ± sd. 5.7 ± 0.7 × 3.3 ± 0.3 μm). Conidiophores borne on aerial mycelia, 50–80 μm tall, unbranched, bearing terminal monophialides, sometimes reduced to single phialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, 30–35 × 3–5 μm, periclinal thickening inconspicuous or absent; aerial microconidia single or forming small false heads on the tips of the monophialides, hyaline, ovoid, reniform, or obovoid with a truncate base, smooth- and thin-walled, aseptate, (5.2–)5.7–8.8(–9.7) × 2.1–3.8 μm (av. ± sd. 7 ± 0.9 × 2.7 ± 0.3 μm). Chlamydospores not observed.
Additional material examined. China, Guangdong Province, Guangzhou city, from species of Orchidaceae, Mar. 2011, Y.Y. Su, LC13607; ibid., LC13608.
Notes — The genus Bisifusarium was established to accommodate several fusarioid species previously included in the F. dimerum species complex, with B. dimerum as type species (Lombard et al. 2015). Prior to this study eight species were known from the genus (Lombard et al. 2015, Sun et al. 2017). Bisifusarium aseptatum is distinct from other Bisifusarium species in producing unicellular sporodochial conidia (Lombard et al. 2015).
Neocosmospora E.F. Sm., U.S.D.A. Div. Veg. Pathol. Bull. 17: 45. 1899
Neocosmospora lithocarpi M.M. Wang & L. Cai, sp. nov. — MycoBank MB 842163; Fig. 22
Fig. 22.
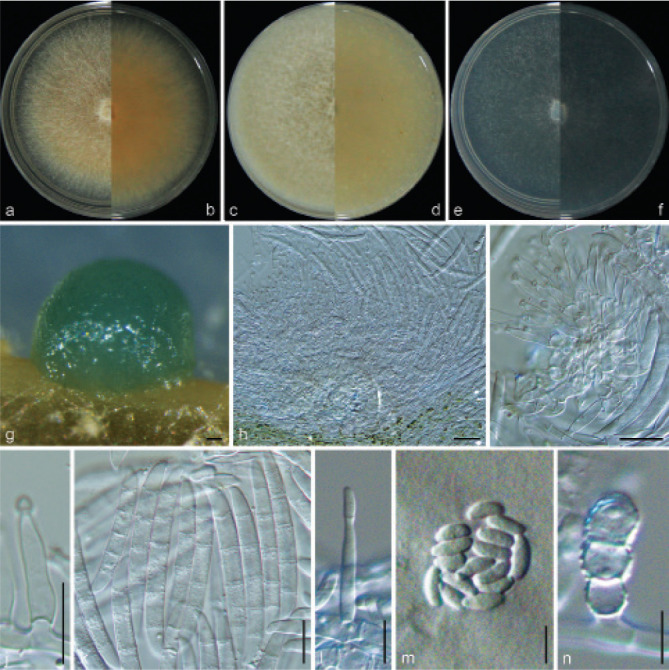
Neocosmospora lithocarpi (ex-type culture LC1113). a–b. Colony on PDA: a. surface of colony on PDA after 7 d at 25 °C; b. reverse of colony on PDA; c–d. colony on OA: c. surface of colony on OA after 7 d at 25 °C; d. reverse of colony on OA; e–f. colony on SNA: e. surface of colony on SNA after 7 d at 25 °C; f. reverse of colony on SNA; g. sporodochium on carnation leaves; h–i. conidiophores and phialides on sporodochia; j. phialide on sporodochia; k. sporodochial conidia (macroconidia); l. phialides on aerial mycelium; m. aerial conidia (microconidia); n. chlamydospores. — Scale bars: g = 50 μm; h = 20 μm; i–n = 10 μm.
Etymology. Named after the host genus Lithocarpus, from which the holotype was isolated.
Typus. China, from Lithocarpus glabra, May 2011, W. Sun (HMAS 351586, holotype designated here, dried culture on SNA with carnation leaves; culture ex-type CGMCC 3.20827 = LC1113).
Colonies on PDA grown in the dark reaching 5.7–5.9 cm diam after 7 d at 25 °C, flat, aerial mycelia dense, colony margin filamentous to erose, filiform, surface white to greyish yellow (4B3) in the centre, white at the margin; reverse greyish orange (5B3) in the centre, white at the margin. Colonies on OA grown in the dark reaching 5.7–5.9 cm diam after 7 d at 25 °C, flat, aerial mycelia dense, colony margin entire, surface white to yellowish grey (4B2) in the centre, white at the margin; reverse yellowish grey (4B2) in the centre, white at the margin. Colonies on SNA grown in the dark reaching 5.2–5.5 cm diam after 7 d at 25 °C, flat, aerial mycelia scant, colony margin erose, white; reverse white. Pigment and odour absent. Sporodochia opaline green (25C6), formed abundantly on carnation leaves. Conidiophores in sporodochia verticillately branched and densely packed; sporodochial phialides subulate to subcylindrical, 9.9–23.3 × 2.8–6.3 μm (av. ± sd. 14.4 ± 2.7 × 4.0 ± 0.8 μm), smooth- and thin-walled, showing a reduced and flared collarette. Sporodochial macroconidia falcate, with a blunt apical and basal cell, 5-septate, hyaline, smooth- and thin-walled, 32.1–57.8 × 3.9–8.1 μm (av. ± sd. 49.6 ± 4.8 × 5.3 ± 0.9 μm). Conidiophores borne on aerial mycelia, 20–60 μm tall, unbranched or sparingly branched, bearing terminal or intercalary monophialides, often reduced to single phialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, 20–45 × 2–3 μm; aerial microconidia forming small false heads on the tips of the monophialides, hyaline, ellipsoid to falcate, smooth- and thin-walled, 0–1-septate; aseptate conidia: (7–)8.8–11.1(–12) × 3.5–3.9 μm (av. ± sd. 9.9 ± 1 × 3.7 ± 0.2 μm); 1-septate conidia: (12–)12.5–23.8(–24) × 3.6–7 μm (av. ± sd. 16.3 ± 2.8 × 5 ± 0.7 μm). Chlamydospores abundant, terminal, ellipsoid, rough, thick-walled, hyaline, aseptate, 6.1–8.7 × 5.3–7.3 μm (av. ± sd. 6.8 ± 0.9 × 5.8 ± 0.7 μm).
Additional material examined. China, from Lithocarpus glabra, May 2011, W. Sun, LC13831; ibid., LC13832.
Notes — Neocosmospora lithocarpi is phylogenetically closely related to N. ambrosia, N. euwallaceae, N. kuroshio, N. oligoseptata, and N. pseudensiformis (Fig. 10). Morphologically, this species is distinguished in the shape, septum number and length of its sporodochial macroconidia (falcate, 5-septate in N. lithocarpi, vs irregularly clavate and swollen conidia present, 3- or 5-septate in N. ambrosia, N. euwallaceae, N. kuroshio, N. oligoseptata; 5-septate, 32.1–57.8 μm in N. lithocarpi, vs 2–8-septate, 49–63 μm in N. pseudensiformis) (Nalim et al. 2011, Aoki et al. 2018, Na et al. 2018, Sandoval-Denis et al. 2019).
DISCUSSION
In this study, 259 species belonging to four well-supported genera, including 12 new species were analysed using a combined tef1-rpb1-rpb2 multi-locus phylogeny (Fig. 1). Within Fusarium, 196 species are categorised in nine species complexes, one of these complexes here renamed as the F. falsibabinda complex (previously incorrectly recognised as the F. babinda species complex) (Fig. 1, 2). One and four distinct clades in the FLSC (Fig. 5) and FOSC (Fig.6), respectively, were not described, awaiting more data for species delimitation. Seven loci were employed in this study, i.e., ITS, IGS, tef1, cam, rpb1, rpb2 and tub2 (Table 1).Among these seven loci, ITS failed to resolve any species in Fusarium, but recognized B. aseptatum, B. penzigii and B. tonghuanum from other known species in Bisifusarium. The IGS locus was amplified for the FOSC members, but its phylogenetic topology showed significant conflict with other loci. The rpb2 locus appeared to be most effective in species recognition in several Fusarium complexes, e.g., the FFSC (Supplementary Fig. S2d), the FTSC (Supplementary Fig. S3d), and Neocosmospora (Supplementary Fig. S5c), followed by tef1 (effective in the FOSC). The rpb1 locus showed the best species recognition in the FNISSC (Supplementary Fig. S1b).
Employing morphological characters, multi-locus phylogenies and ecological preferences, 356 fusarioid isolates from China were identified to 72 species belonging to three genera (1 species with 3 isolates in Bisifusarium, 60 species with 321 isolates in Fusarium and 11 species with 32 isolates in Neocosmospora). Most of the previous studies on Fusarium in China focused on species associated with agricultural and cash crops, e.g., maize, rice, wheat, pepper, and tobacco (Yu 1955, Tai 1979, Wang et al. 2013a, b, Zhang et al. 2014a, b), and insects (Bai & Chen 1991), with more than 250 species, subspecies, varieties, and formae specialis hitherto reported. Based on the current taxonomy, these records correlate to 87 species, as summarised in Table 4. Our results present a significant step towards understanding the species diversity and distribution of Fusarium in China (Table 1), which increased the number of species to 114 (11 new species and 16 new records from China (23.7 % increase)).
Table 4.
Species of fusarioid genera occurring in China, with information of their habitats, hosts and references.
| Current name | Name of taxon | Host genera/habitats | Recorded reference/database |
|---|---|---|---|
| A. rigidiuscula | F. decemcellulare, F. rigidiusculum | Coccids; Dianthus, Garuga, Idiocerus, Passiflora, Salix, Oxytropis | Bai & Chen (1991); https://nmdc.cn/fungarium/fungi/chinadirectories |
| B. aseptatum | B. aseptatum | Orchidaceae | this study |
| B. delphinoides | F. delphinoides | Human | https://nmdc.cn/fungarium/fungi/chinadirectories |
| B. dimerum | F. dimerum | Soil; Citrus, Musa | Yu (1955), Tai (1979); https://nmdc.cn/fungarium/fungi/chinadirectories |
| C. cavisperma | F. cavispermum | Pinus | Yu (1955) |
| F. acaciae-mearnsii | F. acaciae-mearnsii | Paederia | this study |
| F. acuminatum | F. acuminatum, F. caudatum, F. scirpi var. acuminatum | Soil; Acer, Brassica, Capsicum, Crotalaria, Cucumis, Eleocharis, Feijoa, Fritillaria, Glycine, Gossypium, Hordeum, Hylotelephium, Ipomoea, Prunus, Solanum, Triticum | Yu (1955), Tai (1979), this study; https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. acutatum | F. acutatum | Triticum | https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. alpinum | F. alpinum | Fabaceae | this study |
| F. ananatum | F. ananatum | Ananas | https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. anguioides | F. anguioides | Alocasia, Cordyline, Gossypium, Ipomoea, Pisum, Solanum, Vicia Malus, Musa, Olea, Oryza, Sorghum, Vitis, Zea; unidentified mushroom | Yu (1955), this study |
| F. annulatum | F. annulatum | Submerged wood; human; bamboo, Capsicum, Chamaerops, Glycine, Lithocarpus, | https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. annuum | F. annuum | Capsicum | Yu (1955); https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. anthophilum | F. anthophilum, F. moniliforme var. anthophilum | Soil; Gossypium, Oryza, Vicia | Tai (1979); https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. aquatilium | F. aquatilium | Water | this study |
| F. arcuatisporum | F. arcuatisporum | Soil; Brassica, Nelumbo, Oryza, Panicum, Paspalum, Poa | Wang et al. (2019), this study |
| F. armeniacum | F. armeniacum | Unidentified grass | this study |
| F. arthrosporioides | F. arthrosporioides | Soil; Oryza, Vicia | Yu (1955); https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. asiaticum | F. asiaticum | Musa, Triticum | this study; https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. avenaceum | F. avenaceum, F. avenaceum var. fabae, F. avenaceum var. herbarum, F. avenaceum f. fabae, F. avenaceum f. fabalis, F. avenaceum f. fabarum | Air, faeces, soil; Allium, Atractylodes, Avena, Bidens, Brassica, Camellia, Chrysanthemum, Cicer, Citrus, Codonopsis, Coix, Cucumis, Cucurbita, Daucus, Dianthus, Dolichus, Equisetum, Fritillaria, Gentiana, Glycine, Gossypium, Halenia, Hordeum, Juglans, Lathyrus, Lycopersicum, Malus, Oryza, Panax, Papaver, Paspalum, Pinus, Pisum, Plantago, Prunus, Rosa, Salvia, Setaria, Solanum, Sorghum, Trifolium, Triticum, Vicia, Vitis, Zea | Yu (1955), Tai (1979), this study; https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. bactridioides | F. bactridioides | Cucumis, Narcissus, Pinus | Yu (1955) |
| F. bambusacearum | F. bambusacearum | Bamboo | this study |
| F. bambusicola | F. bambusicola | Sinocalamus | Tai (1979) |
| F. brachygibbosum | F. brachygibbosum | Zea | https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. caeruleum | F. coeruleum, F. solani var. caeruleum, F. solani var. coeruleum | Soil; Abies, Aconitum, Actinidia, Aloe, Amorphopha, Arachis, Artocarpus, Astragalus, Atrachylis, Atractylodes, Brassica, Camellia, Canna, Capsicum, Castanea, Chrysomphalus, Citrus, Cucumis, Cuminum, Cyamopsis, Cymbidium, Dendrobium, Dianthus, Dioscorea, Ditylenchus, Eleocharis, Ephedra, Epiphyllum, Eria, Fragaria, Fritillaria, Glycine, Gossypium, Haloxylon, Hcarites, Heleocharis, Helianthus, Heliothis,Helwingia, Heterodera, Hevea, Ipomoea, Jatropha, Juglans, Lablab, Lentinus, Ligusticum, Lilium, Lotus, Luffa, Lycium, Malus, Mangifera, Medicago, Momordica, Morus, Musa, Nicotiana, Nopalxochia, Onobrychis, Oryza, Paeonia, Panax, Paphiopedilum, Passiflora, Persica, Phaius, Phalaenopsis, Phaseolus, Phyllostachys, Pinus, Pisum, Pittosporum, Punica, Rabdosia, Ricinus, Robinia, Salvia, Schisandra, Simmondsia, Solanum, Triticum, Vicia, Vigna, Zanthoxylum, Zea; coccids, human | Yu (1955), Tai (1979), Bai & Chen (1991), this study; https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. camptoceras | F. camptoceras | Soil; Cinnamomum, Clerodentron, Hordeum, Malus, Momordica, Musa, Phaseolus, Triticum, Vigna, Zea | this study; https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. cassiae | F. cassiae | Coffea | this study |
| F. chlamydosporum | F. chlamydosporum, F. fusarioides | Aspidiotus, Auricularia, Glycine, Hordeum, Jacaranda, Juglans, Musa, Parlatoria, Triticum, Zea | https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. chongqingense | F. chongqingense | Bothrocaryum | this study |
| F. citri | F. citri | Amygdalus, Capsicum, Castanopsis, Citrus, Musa, Smilax | Wang et al. (2019), this study |
| F. commune | F. commune | Eleocharis, Musa, Oryza, Vigna | this study; https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. compactum | F. compactum, F. scirpi var. compactum | Soil; Poa, Setaria | this study; https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. concentricum | F. concentricum | Aglaonema, Hedera, Lablab, Maianthemum, Musa, Reineckia, Vitis | this study; https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. concolor | F. concolor, F. polyphialidicum | Soil; Anacardium, Carica, Citrus, Ehretia, Gossypium, Hemerocallis, Hordeum, Lycium, Lygodium, Malva, Rhynchites, Setaria, Triticum, Vicia, Zea | Yu (1955), this study; https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. cugenangense | F. cugenangense | Poa, Smilax, Solanum, Zingiber | this study |
| F. culmorum | F. culmorum | Asparagus, Avena, Beta, Camellia, Cucumis, Cucurbita, Daucus, Dianthu, Diaphorina, Glycine, Helianthus, Hordeum, Ipomoea, Linum, Lycopersicum, Oryza, Pinus, Secale, Setaria, Solanum, Sorghum, Triticum, Vicia, Zea | Yu (1955), Tai (1979); https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. diversisporum | F. diversisporum | Setaria, Zea | Yu (1955) |
| F. duoseptatum | F. duoseptatum | Musa | this study |
| F. elaeagni | F. elaeagni | Elaeagnus | this study |
| F. elaeidis | F. elaeidis | Caryota | this study |
| F. equiseti | F. equiseti | Faeces, soil; aphids, cicadas, coccids; Aphis, Atractylodes, Beta, Calotropis, Capsicum, Castanea, Citrus, Coix, Cucumis, Cucurbita, Cyamopsis, Cymbidium, Dendrobium, Dendrolimus, Eleocharis, Glycine, Gossypium, Helianthus, Henosepilachua, Hordeum, Icerya, Laccifer, Medicago, Momordica, Nicotiana, Oryza, Phaseolus, Pieris, Populus, Prunus, Pseudostellaria, Raphanus, Rhynchites, Rosa, Schisandra, Setaria, Simmondsia, Solanum, Spinacia, Triticum, Vicia, Vigna, Zea | Yu (1955), Tai (1979), Bai & Chen (1991), this study; https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. flocciferum | F. flocciferum | Soil | https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. fujikuroi | F. fujikuroi, F. fujikuroi var. subglutinans, F. moniliforme, F. moniliforme f. subglutinans, F. moniliforme var. hangzhouense, F. moniliforme var. intermedium, F. moniliforme var. minus, F. moniliforme var. minus, F. moniliforme var. subglutinans, F. moniliforme var. subglutinans | Faeces, putty, slug moths, soil, submerged wood; human, aphids, arachids, cicadas, coccids; Aleurocanthus, Allium, Amygdalus, Anoplophora, Areca, Asparagus, Avena, Bamboo, Bombyx, Brassica, Camellia, Capsicum, Cedus, Celosia, Ceroplastes, Chilo, Chlorops, Citrullus, Citrus, Cnaphalocrocis, Cocos, Coffea, Coix, Cucumis, Cucurbita, Cymbidium, Dianthus, Dolichos Elaeis, Eleocharis, Ephedra, Erigeron, Ficus, Gladiolus, Glycine, Gossypium, Helianthus, Hisbiscus, Hordeum, Hyphantria, Icerya, Inazuma, Ipomoea, Jasminum, Jatropha, Juglans, Laccifer, Lilium, Lycium, Lycopersicon, Lycopersicum, Lygodium, Malus, Mangifera, Melia, Musa, Mythimna, Nephotettix, Nilaparvata, Oryza, Ostrinia, Paeonia, Panax, Panicum, Papilio, Parlatoria, Parnara, Paulowine, Phaseolus, Phragmites, Pieris, Pinus, Pleurotus, Populus, Procera, Pseudaonidia, Raphanus, Rhododendron, Rhynchites, Ricinus, Rosa, Saccharum, Sanseviera, Saperda, Scirpophaga, Sesamia, Setaria, Simmondsia, Solanum, Sorghum, Spinacia, Taiwania, Taxus, Trifolium, Triticum, Tulipa, Unaspis, Vicia, Vigna, Zanthoxylum, Zea | Yu (1955), Tai (1979), Bai & Chen (1991), this study; https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. graminearum | F. graminearum, F. zeae | Air, faeces, soil; coccids, psyllids; Agropyron, Allium, Astragalus, Avena, Brassica, Capsicum, Carthamus, Castanea, Coix, Coriolus, Coronilla, Cucumis, Dactylis, Eleocharis, Glycine, Gossypium, Helianthus, Hordeum, Linum, Lolium, Lotus, Lycopersicum, Malus, Medicago, Melilotus, Melissitus, Onobrychis, Oryza, Phalaenopsis, Phaseolus, Pisum, S Populus, Roegneria, ecale, Setaria, Solanum, Sorghum, Spiraea, Triticum, Vicia, Vigna, Zea | Yu (1955), Tai (1979), Bai & Chen (1991), this study; https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. graminum | F. graminum | Paspalum, Vicia | Yu (1955); https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. grosmichelii | F. grosmichelii | Chamaerops, Musa, Oryza | this study |
| F. guilinense | F. guilinense | Musa | Wang et al. (2019), this study |
| F. hainanense | F. hainanense | Musa, Oryza | Wang et al. (2019), this study |
| F. hechiense | F. hechiense | Musa | this study |
| F. heterosporum | F. heterosporum | Soil; Phyllostachys | https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. humuli | F. humuli | Cedrela, Chimonanthus, Coriaria, Humulus, Ligustrum, Liquidambar, Megathyrsus, Musa, Osmanthus, Paederia, Rosa, Viburnum, Vinca | Wang et al. (2019), this study |
| F. incarnatum | F. incarnatum, F. semitectum | Soil; Juglans, Lycopersicum, Miscanthus, Pennisetum, Phragmites, Schoenoplectus, Solanum | Tai (1979), this study; https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. ipomoeae | F. ipomoeae | Soil, submerged wood; Agrostis, bamboo, Capsicum, Hibiscus, Hosta, Ipomoea, Lagenaria, Musa, Oryza, Rhododendron, Solanum, Vinca | Wang et al. (2019), this study |
| F. iranicum | F. iranicum | Lithocarpus | this study |
| F. irregulare | F. irregulare | Bamboo, Digitaria, Lithocarpus, Vigna | Wang et al. (2019), this study |
| F. kyushuense | F. kyushuense | Submerged wood; Chamaedaphne, Lithocarpus, Musa | this study; https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. lacertarum | F. lacertarum | Capsicum | Wang et al. (2019), this study |
| F. lactis | F. lactis | Pisum | Yu (1955) |
| F. langsethiae | F. langsethiae | Unknown | https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. lateritium | F. baccata, F. baccata var. moricola, F. lateritium, F. lateritium f. sp. celosiae, F. lateritium f. mori, F. lateritium var. mori | Soil, wood; coccids; Allium, Aralia, Brevicoryne, Callistephus, Capsicum, Celosia, Chlorops, Chrysomphalus, Cicadella, Citrus, Dendrobium, Diaspiaiotus, Eleutherococcus, Garuga, Ginkgo, Heterodera, Hordeum, Ilex, Laccifer, lcerya, Malus, Malva, Morus, Musa, Phynchites, Pinus, Prinsepia, Prunus, Pulvinaria, Pyrus, Taxus, Triticum | Yu (1955), Tai (1979), Bai & Chen (1991), this study; https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. longipes | F. equiseti var. longipes, F. longipes | Paspalum, Phaseolus | this study; https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. luffae | F. luffae | Humulus, Luffa, Oryza | Wang et al. (2019), this study |
| F. lumajangense | F. lumajangense | Arenga, Musa | this study |
| F. madaense | F. madaense | Oryza | this study |
| F. mangiferae | F. mangiferae | Mangifera | https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. meridionale | F. meridionale | Carbonatite; Coffea, Musa | this study |
| F. miscanthi | F. miscanthi | Water | this study |
| F. mundagurra | F. mundagurra | Paspalum | this study |
| F. nanum | F. nanum | Musa | this study |
| F. napiforme | F. napiforme | Juglans | https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. nepalense | F. nepalense | Camellia, Musa | this study |
| F. nirenbergiae | F. nirenbergiae | Caryota, Musa, Setaria | this study |
| F. odoratissimum | F. odoratissimum | Musa | this study |
| F. oxysporum | F. bulbigenum, F. bulbigenum var. batatas, F. bulbigenum var. lycopersici, F. bulbigenum var. niveam, F. bulbigenum var. tracheiphilum, F. conglutinans, F. conglutinans var. batae F. conglutinans var. callistephi, F. dianthi, F. lini, F. orthoceras, F. orthoceras var. longius, F. orthoceras var. longius, F. orthoceras var. pisi, F. oxysporum, F. oxysporum f. apii, F. oxysporum f. betae, F. oxysporum f. callistephi, F. oxysporum f. ciceris, F. oxysporum f. conglutinans, F. oxysporum f. cubense, F. oxysporum f. cucumerinum, F. oxysporum f. cyclaminis, F. oxysporum f. dianthi, F. oxysporum f. gladioli, F. oxysporum f. lini, F. oxysporum f. lupini, F. oxysporum f. lycopersici, F. oxysporum f. medicaginis, F. oxysporum f. melonis, F. oxysporum f. niveum, F. oxysporum f. perniciosum, F. oxysporum f. phaseoli, F. oxysporum f. pisi, F. oxysporum f. spinaciae, F. oxysporum f. tracheiphilum, F. oxysporum f. tuberosi, F. oxysporum f. vasinfectum, F. oxysporum f.sp. batatas, F. oxysporum f.sp. benincasae, F. oxysporum f.sp. betae, F. oxysporum f.sp. conglutinans, F. oxysporum f.sp. cubense, F. oxysporum f.sp. cucumerinum, F. oxysporum f.sp. cumini, F. oxysporum f.sp. dioscoreae, F. oxysporum f.sp. fabae, F. oxysporum f.sp. glycines, F. oxysporum f.sp. lagenariae, F. oxysporum f.sp. lini, F. oxysporum f.sp. lycopersici, F. oxysporum f.sp. magnoliae, F. oxysporum f.sp. melonis, F. oxysporum f.sp. momordicae, F. oxysporum f.sp. mormodicae, F. oxysporum f.sp. narcissi, F. oxysporum f.sp. nelumbicola, F. oxysporum f.sp. niveum, F. oxysporum f.sp. phaseoli, F. oxysporum f.sp. pisi, F. oxysporum f.sp. vasinfectum, F. oxysporum f. fabae, F. oxysporum var. aurantiacum, F. oxysporum var. cubense, F. oxysporum var. gladioli, F. oxysporum var. orthoceras, F. vasinfectum, F. vasinfectum var. sesami | Faeces, gluten, soil, wood; Acacia, Allium, Amygdalus, Apium, Apocynum, Arachis, Asparagus, Atractylodes, Avena, Benincasa, Beta, Brassica, Callistephus, Camptotheca, Capsicum, Cicer, Citrullus, Citrus, Coriolus, Cucumis, Cucurbita, Cyclamen, Datura, Delonix, Dendranthema, Dianthus, Dioscorea, Fragaria, Gladiolus, Glycine, Gossypium, Helianthus, Hibiscus, Iphigenia, Ipomoea, Iris, Jasminum, Larix, Lilium, Linum, Lupinus, Lycium, Lycopersici, Lycopersicon, Lycopersicum, Magnolia, Malus, Medicago, Momordica, Musa, Narcissus, Nelumbo, Onobrychis, Phascolus, Phaseolus, Pinus, Piper, Pisum, Sechium, Sesamum, Setaria, Solanum, Spinacia, Tamarix, Trifolium, Triticum, Vanilla, Vicia, Vitrullus, Zea | Yu (1955), Tai (1979), Bai & Chen (1991), this study; https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. paeoniae | F. paeoniae | Crataegus, Elymus, Paeonia, Populus | this study |
| F. panlongense | F. panlongense | Musa | this study |
| F. paranisikadoi | F. paranisikadoi | Unidentified grass | this study |
| F. pernambucanum | F. pernambucanum | Bamboo, Capsicum, Chamaedorea, Cyperus, Gerbera, Heteropogon, Musa, Panicum, Zea | this study |
| F. poae | F. poae | Soil; Avena, Cucumis, Dianthus, Oryza, Setaria, Triticum, Zea | Yu (1955), this study; https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. proliferatum | F. proliferatum | Human; Celastrus, Citrullus, Cucumis, Dendrobium, Eleocharis, Glycyrrhiza, Juglans, Musa, Oryza, Oxytropis, Solanum, Vitis | Yu (1955), this study; https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. pseudocircinatum | F. pseudocircinatum | Syzygium | this study |
| F. pseudograminearum | F. pseudograminearum | Triticum | https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. redolens | F. oxysporum var. redolens, F. redolens | Soil; arachnids, cicadas, coccids, scarabs, white flies; Acaeia, Alaugium, Aleurocanthus, Allium, Ananas, Anthurium, Arachis, Astragalus, Atractylodes, Auricularia, Avicennia, Bambusoideae, Bauhinia, Benincasa, Brassica, Brevicoryne, Bupleurum, Callistephus, Camellia, Cannabis, Capsicum, Castanea, Catharanthus, Citrullus, Citrus, Coronilla, Cucumis, Cuminum, Cyamopsis, Cymbidium, Dendrobium, Dianthus, Dimocarpus, Ditylenchus, Dracaena, Elacis, Eleocharis, Ephedra, Excoecaria, Fragaria, Fritillaria, Gerbera, Ginkgo, Gladiolus, Glycine, Glycyrrhiza, Gossypium, Gymnospermae, Gynotemma, Haloxylon, Helianthus, Hevea, Hordeum, Jacaranda, Juglans, Juniporus, Lablab, Laccifer, Lagenaria, Larix, Lcerya, Lentinus, Ligusticum, Lilium, Linum, Llgusticum, Luffa, Lycium, Lycoperdon, Lycopersicon, Magnolia, Medicago, Melilotus, Momordica, Murraya, Musa, Myrica, Nelumbo, Nicotiana, Nopalxochia, Onobrychis, Oryza, Oxytropis, Paeonia, Panax, Parlatoria, Pennisetum, Phalaenopsis, Phaseolus, Pinus, Piper, Pisum, Pocockia, Populus, Rabdosia, Raphanus, Rheum, Rhodiola, Rhynchites, Robinia, Saccharum, Saperda, Schisandra, Simmondsia, Solanum, Sorghum, Spinacia, Spiraea, Stevia, Taxus, Trifolium, Triticum, Unaspis, Ustilago, Vernicia, Vicia, Vigna, Vitis, Zea, Zingiber | Yu (1955), Tai (1979), Bai & Chen (1991); https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. reticulatum | F. pallidoroseum, F. semitectum var. majus | Beta, Cucumis, Cucurbita | Yu (1955) |
| F. sacchari | F. sacchari | Arundina, Musa, Poa | this study |
| F. sambucinum | F. pulicaris, F. roseum, F. sambucinum | Soil; bamboo, Ceroplastes, Citrug, Citrus, Dendrobium, Panax, Ricinus, Saccharum, Solanum, Triticum, Zanthoxylum, Zea | Yu (1955), Tai (1979); https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. sarcochroum | F. sarcochroum | Populus | https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. scirpi | F. scirpi | Soil; Avena, Citrus, Gladiolus, Gossypium, Hordeum, Panax, Pinus, Setaria | Yu (1955), Tai (1979); https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. sinense | F. sinense | Triticum | https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. sporotrichioides | F. solani var. martii forma 3, F. sporotrichioides, F. sporotrichiella var. sporotrichioides, F. sporotrichioides var. chlamydosporum | Soil; Arachis, Gossypium, Helianthus, Juglans, Phaseolus, Pisum, Pseudaonidia, Solanum, Zea | Yu (1955); https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. stilboides | F. stilboides | Clausena, Coffea | this study; https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. subglutinans | F. subglutinans | Chilo, Dianthus, Ephedra, Erigeron, Raphanus, Zea | https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. sulawense | F. sulawense | Acalypha, Alocasia, bamboo, Capsicum, Citrus, Colocasia, Ipomoea, Luffa, Musa, Oryza, Smilax, Syngonium | Wang et al. (2019), this study |
| F. tanahbumbuense | F. tanahbumbuense | Digitaria, Oryza | Wang et al. (2019), this study |
| F. temperatum | F. temperatum | Unidentified lichen | this study |
| F. thapsinum | F. thapsinum | Human | https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. tricinctum | F. tricinctum | Dendrobium, Glycine, Hordeum, Hosta, Litsea, Pisum, Setaria, Solanum, Sophora, Triticum, Vicia, Zamia, Zea | Yu (1955), this study; https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. udum | F. udum, F. udum f.sp. crotalariae | Soil, wood; Amygdalus, Asarum, Citrullus, Coriolus, Gossypium, Salix, Schisandra, Solanum | https://nmdc.cn/fungarium/fungi/chinadirectories |
| F. ussurianum | F. ussurianum | Air, carbonatite, soil; bamboo, Musa, Oryza, Podocarpus, Prunus, Rhynchospora | this study |
| F. verticillioides | F. verticillioides | Soil, submerged wood; Anoplophora, bamboo, Brassica, Citrullus, Cucumis, Glycine, human, Hyphantria, Musa, Phaseolus, Physosfegia, Saperda, Solanum, Vigna, Zea | this study; https://nmdc.cn/fungarium/fungi/chinadirectories |
| N. brevis | N. brevis | Submerged wood | this study |
| N. falciformis | N. falciformis | Paspalum, Passiflora, Vitis | this study |
| N. lithocarpus | N. lithocarpus | Lithocarpus | this study |
| N. metavorans | N. metavorans | Submerged wood | this study |
| N. oblonga | N. oblonga | Carbonatite | this study |
| N. paraeumartii | N. paraeumartii | Castanopsis | this study |
| N. petroliphila | N. petroliphila | Lithocarpus | this study |
| N. phaseoli | N. phaseoli | Brassica, Vigna | https://nmdc.cn/fungarium/fungi/chinadirectories |
| N. pseudensiformis | N. pseudensiformis | Passiflora | this study |
| N. silvicola | N. silvicola | Faeces | this study |
| N. solani | F. eumartii, F. javanicum, F. javanicum var. radicicola, F. martii, F. solani, F. solani f. batatus, F. solani f.sp. aleuritidis, F. solani f.sp. fabae, F. solani f.sp. lilii, >F. solani f.sp. pisi, F. solani var. eumartii, F. solani var. javanicum, F. solani var. martii, F. solani var. solani | Compost, faeces, soil; coccids; Allium, Amomum, Amygdalus, Benincasa, Callistephus, Capsicum, Chrysanthemum, Citrullus, Citrus, Colocasia, Cucumis, Dimocarpus, Dioscorea, Glycine, Gossypium, Ipomoea, Lilium, Lycopersicum, Musa, Oryza, Panax, Phaseolus, Pinus, Pisum, Polygatum, Polygonatum, Rehmannia, Robinia, Santalum, Solanum, Vernicia, Vicia, Zingiber | Yu (1955), Tai (1979), this study; https://nmdc.cn/fungarium/fungi/chinadirectories |
| N. stercicola | N. stercicola | Soil | this study |
| R. ventricosum | F. ventricosum | Soil; sawfly pupae; Camellia, Icerya | Bai & Chen (1991); https://nmdc.cn/fungarium/fungi/chinadirectories |
| S. ciliatum | F. ciliatum | Populus | Yu (1955) |
Note: A. = Albonectria, B. = Bisifusarium, C. = Cosmospora, F. = Fusarium, N. = Neocosmospora, R. = Rectifusarium, S. = Scolecofusarium.
In this study, 321 isolates from China were characterised belonging to eight species complexes in Fusarium. The F. fujikuroi complex (FFSC) presented the widest geographic distribution (30 locations) and the second highest number of hosts/ habitats (28 types; 20 plant genera). Members in this complex were well-known for their worldwide distribution, with wide host ranges, diverse habitats, and pathogenicity to many cereals and economically relevant plants (O’Donnell et al. 1998a, 2000b, Leslie & Summerell 2006). Hitherto about 73 species were accepted in this complex, and 17 of them (15 from China, two from the USA), including four novel species and four newly recorded species in China, were reported in this study. The
F. incarnatum-equiseti complex (FIESC) presented the second widest geographic distribution (21 locations) and the highest number of hosts/habitats (50 types; 44 plant genera). This complex is well-known as pathogens of plants and animals, endophytes, and saprobes of various host substrates (Leslie & Summerell 2006, O’Donnell et al. 2009b, Villani et al. 2016). Currently about 38 species have been introduced in the FIESC (O’Donnell et al. 2009b, Wang et al. 2019, Xia et al. 2019). In this study, 15 species in the complex were reported from environmental habitats and 44 plant genera (29 families) in China (Table 1), suggesting a very wide host range of this complex. The F. tricinctum species complex (FTSC) is an important group in Fusarium which encompasses mycotoxin producing species (Leslie & Summerell 2006). Members of this complex are well-known as cereal grain inhabitants (Kulik 2008), smut and mushroom endophytes (Torbati et al. 2019), and saprobes in soil and other environmental habitats (Leslie & Summerell 2006). Previously four species in this complex were recorded from China, i.e., F. acuminatum, F. avenaceum, F. flocciferum, and F. tricinctum from cereals, pumpkin, and winter squash (Yu 1955, Tai 1979, Zhuang 2005, Zhang et al. 2015, Chang et al. 2018, Li et al. 2019). In this study, seven species were identified, including three new species and one species newly recorded from China (F. iranicum).
This study also investigated 66 strains isolated and intercepted at Ningbo Customs from various economically important plants of 12 countries over six years (2012–2017), e.g., cereals, ornamental plants, and fruits (Table 1). These strains were identified as 26 species, including 25 known and one new species (Table 1). Six known species were hereto undetected in China, and two of them, F. curvatum and N. pisi, were previously reported as pathogens of Brassicaceae and Rubiaceae plants and Pisum sativum, respectively (Lombard et al. 2019a, Sandoval-Denis et al. 2019). Interception of these species implies potential threats to biosafety and ecosystem stability of China in international trade.
The taxonomic framework of Fusarium and allied genera has undergone several significant changes since establishment. Despite controversial opinions that exist on the generic boundaries of Fusarium and allied genera, we favour separating Fusarium s.lat. into multiple genera including Albonectria, Bisifusarium, Fusarium, Neocosmospora, and Rectifusarium as proposed by Gräfenhan et al. (2011), Nalim et al. (2011), Schroers et al. (2011), Lombard et al. (2015), and Crous et al. (2021). Approximately 1800 Fusarium and allied species epithets are recorded in the Index Fungorum and MycoBank databases (accessed December 2021). However, presently less than 400 species are accepted and have been studied using multi-locus DNA data from type specimens (Aoki et al. 2014, O’Donnell et al. 2009a, b, Sandoval-Denis et al. 2018a, b, Lombard et al. 2019a, b, Wang et al. 2019, Xia et al. 2019, Crous et al. 2021, Yilmaz et al. 2021). The taxonomic status of many names remains unresolved because of the lack of type specimens and derived sequences, e.g., F. caeruleum. It is noteworthy that there is still an incredibly high number of undescribed Fusarium species, e.g., 256 phylogenetic clades recorded in Fusarium-ID database (http://isolate.fusariumdb.org/blast.php) vs hitherto only about 31 species introduced in the FOSC (Lombard et al. 2019a). Considering the huge number of fusarioid strains/specimens from diverse fungaria around the world, there are undoubtedly many as yet resolved new species. Significant effort is thus required to fully discern the complexity of such a diverse and important fungal group. It is hoped that with the epitypification and neotypification of old names, and description of cryptic species, the classification system of this fungal group would be more stable and less artificial and serve the needs of the users and community impacted by this fungal group in future.
Acknowledgments
This study was financially supported by the National Natural Science Foundation of China (NSFC 32100005, NSFC 31770009, NSFC 31725001), the China Postdoctoral Science Foundation (2020M680721), and Biological Resources Programme, Chinese Academy of Sciences (KFJBRP-009).
Supplementary material
Fifty percent majority rule consensus trees from Bayesian analyses inferred from the tef1 (a), rpb1 (b), and rpb2 (c), showing the phylogenetic relationships of five species complexes within the Fusarium, namely F. concolor (FCOSC), F. falsibabinda (FFBSC), and F. nisikadoi (FNISSC). The Bayesian posterior probabilities (PP > 0.9) and PhyML Bootstrap support values (BS > 50) are displayed at the nodes (PP/ML). The tree was rooted to Fusarium humuli (CQ1039). Ex-type cultures are indicated with ‘T’.
Fifty percent majority rule consensus trees from Bayesian analyses inferred from the tef1 (a), cam (b), rpb1 (c), rpb2 (d), and tub2 (e), showing the phylogenetic relationships of species within the Fusarium fujikuroi species complex (FFSC). The Bayesian posterior probabilities (PP > 0.9) and PhyML Bootstrap support values (BS > 50) are displayed at the nodes (PP/ML). All the trees were rooted to F. nirenbergiae (CBS 744.97). Ex-type cultures are indicated with ‘T’, epi-type with ‘ET’, neotype with ‘NT’.
Fifty percent majority rule consensus trees from Bayesian analyses inferred from the ITS (a), tef1 (b), rpb1 (c), and rpb2 (d) showing the phylogenetic relationships of species within the Fusarium tricinctum species complex (FTSC). The Bayesian posterior probabilities (PP > 0.9) and PhyML Bootstrap support values (BS > 50) are displayed at the nodes (PP/ML). The tree was rooted to F. concolor (NRRL 13994 T). Ex-type cultures are indicated with ‘T’, neotype with ‘NT’.
Fifty percent majority rule consensus trees from Bayesian analyses inferred from the ITS (a), tef1 (b), rpb2 (c), cam (d), and tub2 (e), showing the phylogenetic relationships of species within the Bisifusarium. The Bayesian posterior probabilities (PP > 0.9) and PhyML Bootstrap support values (BS > 50) are displayed at the nodes (PP/ML). All the trees were rooted to Rectifusarium robinianum (CBS 430.91 T). Ex-type cultures are indicated with ‘T’.
Fifty percent majority rule consensus trees from Bayesian analyses inferred from the ITS (a), tef1 (b), and rpb2 (c), showing the phylogenetic relationships of species within the genus Neocosmospora. The Bayesian posterior probabilities (PP > 0.9) and PhyML Bootstrap support values (BS > 50) are displayed at the nodes (PP/ML). The tree was rooted to Geejayessia cicatricum (CBS 125552) and G. atrofusca (NRRL 22316). Ex-type cultures are indicated with ‘T’, neo-type with ‘NT’.
REFERENCES
- Aoki T, Kasson MT, Berger MC, et al. 2018. Fusarium oligoseptatum sp. nov., a mycosymbiont of the ambrosia beetle Euwallacea validus in the Eastern U.S. and typification of F. ambrosium. Fungal Systematics and Evolution 1: 23–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki T, O’Donnell K, Geiser DM. 2014. Systematics of key phytopathogenic Fusarium species: current status and future challenges. Journal of General Plant Pathology 80: 189–201. [Google Scholar]
- Aoki T, O’Donnell K, Scandiani M. 2005. Sudden death syndrome of soybean in South America is caused by four species of Fusarium: Fusarium brasiliense sp. nov., F. cuneirostrum sp. nov., F. tucumaniae and F. virguliforme. Mycoscience 46: 162–183. [Google Scholar]
- Bai FY, Chen QT. 1991. Fusarium species on some insects from China. Acta Mycologica Sinica 10: 120–128. [Google Scholar]
- Booth C. 1971. The genus Fusarium. Commonwealth Mycological Institute, Kew, Surrey, UK. [Google Scholar]
- Chang X, Dai H, Wang D, et al. 2018. Identification of Fusarium species associated with soybean root rot in Sichuan Province, China. European Journal of Plant Pathology 151: 563–577. [Google Scholar]
- Coleman JJ, Rounsley SD, Rodriguez-Carres M, et al. 2009. The genome of Nectria haematococca: contribution of supernumerary chromosomes to gene expansion. PLoS Genetics 5: e1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ, Summerell BA, et al. 2009. Co-occurring species of Teratosphaeria on Eucalyptus. Persoonia 22: 38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Lombard L, Sandoval-Denis M, et al. 2021. Fusarium: more than a node or a foot-shaped basal cell. Studies in Mycology 98: 100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Verkley GJM, Groenewald JZ, et al. 2019. Fungal Biodiversity. In: Westerdijk Laboratory Manual Series 1. Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands. [Google Scholar]
- Cuomo CA, Guldener U, Xu J-R, et al. 2007. The genome sequence of Fusarium graminearum reveals localized diversity and pathogen specialization. Science 317: 1400–1402. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, et al. 2012. jModelTest 2: more models new heuristics and parallel computing. Nature Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins AE, Munkvold GP, Plattner RD, et al. 2002. FUM1 – a gene required for fumonisin biosynthesis but not for maize ear rot and ear infection by Gibberella moniliformis in field tests. Molecular Plant-Microbe Interactions 15: 1157–1164. [DOI] [PubMed] [Google Scholar]
- Fisher NL, Burgess LW, Toussoun TA, et al. 1982. Carnation leaves as a substrate and for preserving cultures of Fusarium species. Phytopathology 72: 151–153. [Google Scholar]
- Gams W, Klamer M, O’Donnell K. 1999. Fusarium miscanthi sp. nov. from Miscanthus litter. Mycologia 91: 263–268. [Google Scholar]
- Geiser DM, Aoki T, Bacon CW, et al. 2013. One fungus, one name: Defining the genus Fusarium in a scientifically robust way that preserves longstanding use. Phytopathology 103: 400–408. [DOI] [PubMed] [Google Scholar]
- Gordon TR, Martyn RD. 1997. The evolutionary biology of Fusarium oxysporum. Annual Review of Phytopathology 35: 111–128. [DOI] [PubMed] [Google Scholar]
- Gordon WL. 1944. The occurrence of Fusarium species in Canada. I. Species of Fusarium isolated from farm samples of cereal seed in Manitoba. Canadian Journal of Research, C 22: 282–286. [Google Scholar]
- Gordon WL. 1952. The occurrence of Fusarium species in Canada. II. Prevalence and taxonomy of Fusarium species in cereal seed. Canadian Journal of Botany 30: 209–251. [Google Scholar]
- Gordon WL. 1954a. The occurrence of Fusarium species in Canada. III. Taxonomy of Fusarium species in the seed of vegetable, forage, and miscellaneous crops. Canadian Journal of Botany 32: 576–590. [Google Scholar]
- Gordon WL. 1954b. The occurrence of Fusarium species in Canada. IV. Taxonomy and prevalence of Fusarium species in the soil of cereal plots. Canadian Journal of Botany 32: 622–629. [Google Scholar]
- Gordon WL. 1956a. The occurrence of Fusarium species in Canada. V. Taxonomy and geographic distribution of Fusarium species in soil. Canadian Journal of Botany 34: 833–846. [Google Scholar]
- Gordon WL. 1956b. The taxonomy and habitats of the Fusarium species in Trinidad, B.W.I. Canadian Journal of Botany 34: 847–864. [Google Scholar]
- Gordon WL. 1959. The occurrence of Fusarium species in Canada. VI. Taxonomy and geographic distribution of Fusarium species on plants, insects, and fungi. Canadian Journal of Botany 37: 257–290. [Google Scholar]
- Gordon WL. 1960. The taxonomy and habitats of Fusarium species from tropical and temperate regions. Canadian Journal of Botany 38: 643–658. [Google Scholar]
- Gräfenhan T, Schroers H-J, Nirenberg HI, et al. 2011. An overview of the taxonomy, phylogeny, and typification of nectriaceous fungi in Cosmospora, Acremonium, Fusarium, Stilbella, and Volutella. Studies in Mycology 68: 79–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, et al. 2010. New algorithms and methods to estimate Maximum-Likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Guo LD, Hyde KD, Liew ECY. 2000. Identification of endophytic fungi from Livistona chinensis based on morphology and rDNA sequences. New Phytologist 147: 617–630. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Jacobs-Venter A, Laraba I, Geiser DM, et al. 2018. Molecular systematics of two sister clades, the Fusarium concolor and F. babinda species complexes, and the discovery of a novel microcycle macroconidium-producing species from South Africa. Mycologia 110: 1189–1204. [DOI] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. 2017. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20: 1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornerup A, Wanscher JH. 1978. Methuen handbook of colour. 3rd ed. London, Eyre Methuen. [Google Scholar]
- Kulik T. 2008. Detection of Fusarium tricinctum from cereal grain using PCR assay. Journal of Applied Genetics 49: 305–311. [DOI] [PubMed] [Google Scholar]
- Laurence MH, Summerell BA, Burgess LW, et al. 2011. Fusarium burgessii sp. nov. representing a novel lineage in the genus Fusarium. Fungal Diversity 49: 101–112. [Google Scholar]
- Laurence MH, Summerell BA, Burgess LW, et al. 2014. Genealogical concordance phylogenetic species recognition in the Fusarium oxysporum species complex. Fungal Biology 118: 374–384. [DOI] [PubMed] [Google Scholar]
- Leslie JF, Summerell BA. 2006. The Fusarium laboratory manual. Blackwell Publishing Professional, 2121 State Avenue, Ames, Iowa 50014, USA. [Google Scholar]
- Li YG, Jiang WY, Jiang D, et al. 2019. First report of fruit rot on postharvest pumpkin caused by Fusarium acuminatum in China. Plant Disease 103: 1035. [Google Scholar]
- Link HF. 1809. Observationes in ordines plantarum naturals, Dissetatio I. Magazin der Gesellschaft Naturforschenden Freunde Berlin 3: 3–42. [Google Scholar]
- Liu F, Weir BS, Damm U, et al. 2015. Unravelling Colletotrichum species associated with Camellia: employing ApMat and GS loci to resolve species in the C. gloeosporioides complex. Persoonia 35: 63–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Whelen S, Hall BD. 1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Molecular Biology and Evolution 16: 1799–1808. [DOI] [PubMed] [Google Scholar]
- Lombard L, Sandoval-Denis M, Lamprecht SC, et al. 2019a. Epitypification of Fusarium oxysporum – clearing the taxonomic chaos. Persoonia 43: 1–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L, Van der Merwe NA, Groenewald JZ, et al. 2015. Generic concepts in Nectriaceae. Studies in Mycology 80: 189–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L, Van Doorn R, Crous PW. 2019b. Neotypification of Fusarium chlamydosporum – a reappraisal of a clinically important species complex. Fungal Systematics and Evolution 4: 183–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasas WFO, Nelson PE, Toussoun TA. 1984. Toxigenic Fusarium species: Identity and mycotoxicology. Pennsylvania State University, University Park, Pennsylvania. [Google Scholar]
- Marasas WFO, Nelson PE, Toussoun TA, et al. 1986. Fusarium polyphialidicum, a new species from South Africa. Mycologia 78: 678–682. [Google Scholar]
- Maryani N, Lombard L, Poerba YS, et al. 2019a. Phylogeny and genetic diversity of the banana Fusarium wilt pathogen Fusarium oxysporum f. sp. cubense in the Indonesian centre of origin. Studies in Mycology 92: 155–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryani N, Sandoval-Denis M, Lombard L, et al. 2019b. New endemic Fusarium species hitch-hiking with pathogenic Fusarium strains causing Panama disease in small-holder banana plots in Indonesia. Persoonia 43: 48–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na F, Carrillo JD, Mayorquin JS, et al. 2018. Two novel fungal symbionts Fusarium kuroshium sp. nov. and Graphium kuroshium sp. nov. of Kuroshio shot hole borer (Euwallacea sp. nr. fornicatus) cause Fusarium dieback on woody host species in California. Plant Disease 102: 1154–1164. [DOI] [PubMed] [Google Scholar]
- Nalim FA, Samuels GJ, Wijesundera RL, et al. 2011. New species from the Fusarium solani species complex derived from perithecia and soil in the Old World tropics. Mycologia 103: 1302–1330. [DOI] [PubMed] [Google Scholar]
- Nelson PE, Toussoun TA, Marasas WFO. 1983. Fusarium species: An illustrated Manual for Identification. Pennsylvania State University Press, University Park, Pennsylvania. [Google Scholar]
- Nirenberg HI. 1976. Untersuchungen ber die morphologische und biologische Differenzierung in der Fusarium-Sektion Liseola. Mitteilungen der Biologischen Bundesanstalt f r Land- und Forstwirtschaft (Berlin-Dahlem) 169: 1–117. [Google Scholar]
- Nirenberg HI, Aoki T. 1997. Fusarium nisikadoi, a new species from Japan. Mycoscience 38: 329–333. [Google Scholar]
- Nirenberg HI, O’Donnell K. 1998. New Fusarium species and combinations within the Gibberella fujikuroi species complex. Mycologia 90: 434–458. [Google Scholar]
- Nylander JAA, Wilgenbusch JC, Warren DL, et al. 2008. AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24: 581–583. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Cigelnik E. 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Cigelnik E, Nirenberg HI. 1998a. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia 90: 465–493. [Google Scholar]
- O’Donnell K, Gueidan C, Sink S, et al. 2009a. A two-locus DNA sequence database for typing plant and human pathogens within the Fusarium oxysporum species complex. Fungal Genetics and Biology 46: 936–948. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Kistler HC, Cigelnik E, et al. 1998b. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences of the United States of America 95: 2044–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Kistler HC, Tacke BK, et al. 2000a. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proceedings of the National Academy of Sciences of the United States of America 95: 7905–7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, McCormick SP, Busman M, et al. 2018. Marasas, et al. 1984 ‘Toxigenic Fusarium Species: Identity and Mycotoxicology’ revisited. Mycologia 110: 1058–1080. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Nirenberg HI, Aoki T, et al. 2000b. A multigene phylogeny of the Gibberella fujikuroi species complex: detection of additional phylogenetically distinct species. Mycoscience 41: 61–78. [Google Scholar]
- O’Donnell K, Rooney AP, Proctor RH, et al. 2013. Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genetics and Biology 52: 20–31. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Sutton DA, Fothergill A, et al. 2008. Molecular phylogenetic diversity, multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani species complex. Journal of Clinical Microbiology 46: 2477–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Sutton DA, Rinaldi MG, et al. 2009b. Novel multilocus sequence typing scheme reveals high genetic diversity of human pathogenic members of the Fusarium incarnatum-F. equiseti and F. chlamydosporum species complexes within the United States. Journal of Clinical Microbiology 47: 3851–3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Sutton DA, Rinaldi MG, et al. 2010. An Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. Journal of Clinical Microbiology 48: 3708–3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. 2008. jModelTest: Phylogenetic Model Averaging. Molecular Biology and Evolution 25: 1253–1256. [DOI] [PubMed] [Google Scholar]
- Wang MM. et al.: Fusarioid species in China Reeb V, Lutzoni F, Roux C. 2004. Contribution of RPB2 to multilocus phylogenetic studies of the euascomycetes (Pezizomycotina, Fungi) with special emphasis on the lichen-forming Acarosporaceae and evolution of polyspory. Molecular Phylogenetics and Evolution 32: 1036–1060. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van der Mark P, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval-Denis M, Guarnaccia V, Polizzi G, et al. 2018a.. Symptomatic Citrus trees reveal a new pathogenic lineage in Fusarium and two new Neocosmospora species. Persoonia 40: 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval-Denis M, Lombard L, Crous PW. 2019. Back to the roots: a reappraisal of Neocosmospora. Persoonia 43: 90–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval-Denis M, Swart WJ, Crous PW. 2018b. New Fusarium species from the Kruger National Park, South Africa. Mycokeys 34: 63–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroers H-J, Gräfenhan T, Nirenberg HI, et al. 2011. A revision of Cyanonectria and Geejayessia gen. nov., and related species with Fusarium-like anamorphs. Studies in Mycology 68: 115–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovgaard K, Nirenberg HI, O’Donnell K, et al. 2001. Evolution of Fusarium oxysporum f. sp. vasinfectum races inferred from multigene genealogies. Phytopathology 91: 1231–1237. [DOI] [PubMed] [Google Scholar]
- Snyder WC, Hansen HN. 1940. The species concept in Fusarium. American Journal of Botany 27: 64–67. [Google Scholar]
- Snyder WC, Hansen HN. 1941. The species concept in Fusarium with reference to section Martiella. American Journal of Botany 28: 738–742. [Google Scholar]
- Snyder WC, Hansen HN. 1945. The species concept in Fusarium with reference to Discolour and other sections. American Journal of Botany 32: 657–666. [Google Scholar]
- Snyder WC, Hansen HN. 1947. Advantages of natural media and environments in the culture of fungi. Phytopathology 37: 420–421. [PubMed] [Google Scholar]
- Snyder WC, Hansen HN. 1954. Variation and speciation in the genus Fusarium. Annals of the New York Academy of Sciences 60: 16–23. [DOI] [PubMed] [Google Scholar]
- Summerell BA, Rugg CA, Burgess LW. 1995. Characterization of Fusarium babinda sp. nov. Mycological Research 99: 1345–1348. [Google Scholar]
- Sun BD, Zhou YG, Chen A-J. 2017. Bisifusarium tonghuanum (Nectriaceae), a novel species of Fusarium-like fungi from two desert oasis plants. Phytotaxa 317: 123–129. [Google Scholar]
- Tai FL. 1979. Sylloge Fungorum Sinicorum. Science Press, Academia Sinica, Peking. [Google Scholar]
- Torbati M, Arzanlou M, Sandoval-Denis M, et al. 2019. Multigene phylogeny reveals new fungicolous species in the Fusarium tricinctum species complex and novel hosts in the genus Fusarium from Iran. Mycological Progress 18: 119–133. [Google Scholar]
- Van der Does C, Duyvesteijn RGE, Goltstein PM, et al. 2008. Expression of effector gene SIX1 of Fusarium oxysporum requires living plant cells. Fungal Genetics and Biology 45: 1257–1264. [DOI] [PubMed] [Google Scholar]
- Van Hove F, Waalwijk C, Logrieco A, et al. 2011. Gibberella musae (Fusarium musae) sp. nov., a recently discovered species from banana is sister to F. verticillioides. Mycologia 103: 570–585. [DOI] [PubMed] [Google Scholar]
- Villani A, Moretti A, De Saeger S, et al. 2016. A polyphasic approach for characterization of a collection of cereal isolates of the Fusarium incarnatumequiseti species complex. International Journal of Food Microbiology 234: 24–35. [DOI] [PubMed] [Google Scholar]
- Wang HC, Wang MS, Xia HQ, et al. 2013a. First report of Fusarium wilt of tobacco caused by Fusarium kyushuense in China. Plant Disease 97: 424. [DOI] [PubMed] [Google Scholar]
- Wang JH, Feng ZH, Han Z, et al. 2013b. First report of pepper fruit rot caused by Fusarium concentricum in China. Plant Disease 97: 1657. [DOI] [PubMed] [Google Scholar]
- Wang MM, Chen Q, Diao YZ, et al. 2019. Fusarium incarnatum-equiseti complex from China. Persoonia 43: 70–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, et al. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al. (eds), PCR protocols: A guide to the methods and applications: 315–322. New York, NY, Academic Press. [Google Scholar]
- Wollenweber HW, Reinking OA. 1935. Die Fusarien, ihre Beschreibung, Schadwirkung und Bekampfung. Verlag Paul Parey, Berlin, Germany. [Google Scholar]
- Xia JW, Sandoval-Denis M, Crous PW, et al. 2019. Numbers to names – restyling the Fusarium incarnatum-equiseti species complex. Persoonia 43: 186–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz M, Sandoval-Denis M, Lombard L, et al. 2021. Redefining species limits in the Fusarium fujikuroi species complex. Persoonia 46: 129–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu DF. 1955. A preliminary list of Fusaria in China. Acta Phytopathologica Sinica 1: 1–18. [Google Scholar]
- Zhang H, Luo W, Pan Y, et al. 2014a. First report of Fusarium maize ear rot caused by Fusarium meridionale in China. Plant Disease 98: 1156. [DOI] [PubMed] [Google Scholar]
- Zhang H, Luo W, Pan Y, et al. 2014b. First report of Fusarium ear rot of maize caused by Fusarium andiyazi in China. Plant Disease 98: 1428. [DOI] [PubMed] [Google Scholar]
- Zhang K, Su YY, Cai L. 2013. An optimized protocol of single spore isolation for fungi. Cryptogamie, Mycologie 34: 349–356. [Google Scholar]
- Zhang XX, Sun HY, Shen CM, et al. 2015. Survey of Fusarium spp. causing wheat crown rot in major winter wheat growing regions of China. Plant Disease 99: 1610–1615. [DOI] [PubMed] [Google Scholar]
- Zhang ZF, Liu F, Zhou X, et al. 2017. Culturable mycobiota from Karst caves in China, with descriptions of 20 new species. Persoonia 39: 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, O’Donnell K, Aoki T, et al. 2016. Two novel Fusarium species that cause canker disease of prickly ash (Zanthoxylum bungeanum) in northern China form a novel clade with Fusarium torreyae. Mycologia 108: 668–681. [DOI] [PubMed] [Google Scholar]
- Zhuang WY. 2005. Fungi of northwestern China. Mycotaxon, Ltd., Ithaca, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fifty percent majority rule consensus trees from Bayesian analyses inferred from the tef1 (a), rpb1 (b), and rpb2 (c), showing the phylogenetic relationships of five species complexes within the Fusarium, namely F. concolor (FCOSC), F. falsibabinda (FFBSC), and F. nisikadoi (FNISSC). The Bayesian posterior probabilities (PP > 0.9) and PhyML Bootstrap support values (BS > 50) are displayed at the nodes (PP/ML). The tree was rooted to Fusarium humuli (CQ1039). Ex-type cultures are indicated with ‘T’.
Fifty percent majority rule consensus trees from Bayesian analyses inferred from the tef1 (a), cam (b), rpb1 (c), rpb2 (d), and tub2 (e), showing the phylogenetic relationships of species within the Fusarium fujikuroi species complex (FFSC). The Bayesian posterior probabilities (PP > 0.9) and PhyML Bootstrap support values (BS > 50) are displayed at the nodes (PP/ML). All the trees were rooted to F. nirenbergiae (CBS 744.97). Ex-type cultures are indicated with ‘T’, epi-type with ‘ET’, neotype with ‘NT’.
Fifty percent majority rule consensus trees from Bayesian analyses inferred from the ITS (a), tef1 (b), rpb1 (c), and rpb2 (d) showing the phylogenetic relationships of species within the Fusarium tricinctum species complex (FTSC). The Bayesian posterior probabilities (PP > 0.9) and PhyML Bootstrap support values (BS > 50) are displayed at the nodes (PP/ML). The tree was rooted to F. concolor (NRRL 13994 T). Ex-type cultures are indicated with ‘T’, neotype with ‘NT’.
Fifty percent majority rule consensus trees from Bayesian analyses inferred from the ITS (a), tef1 (b), rpb2 (c), cam (d), and tub2 (e), showing the phylogenetic relationships of species within the Bisifusarium. The Bayesian posterior probabilities (PP > 0.9) and PhyML Bootstrap support values (BS > 50) are displayed at the nodes (PP/ML). All the trees were rooted to Rectifusarium robinianum (CBS 430.91 T). Ex-type cultures are indicated with ‘T’.
Fifty percent majority rule consensus trees from Bayesian analyses inferred from the ITS (a), tef1 (b), and rpb2 (c), showing the phylogenetic relationships of species within the genus Neocosmospora. The Bayesian posterior probabilities (PP > 0.9) and PhyML Bootstrap support values (BS > 50) are displayed at the nodes (PP/ML). The tree was rooted to Geejayessia cicatricum (CBS 125552) and G. atrofusca (NRRL 22316). Ex-type cultures are indicated with ‘T’, neo-type with ‘NT’.


