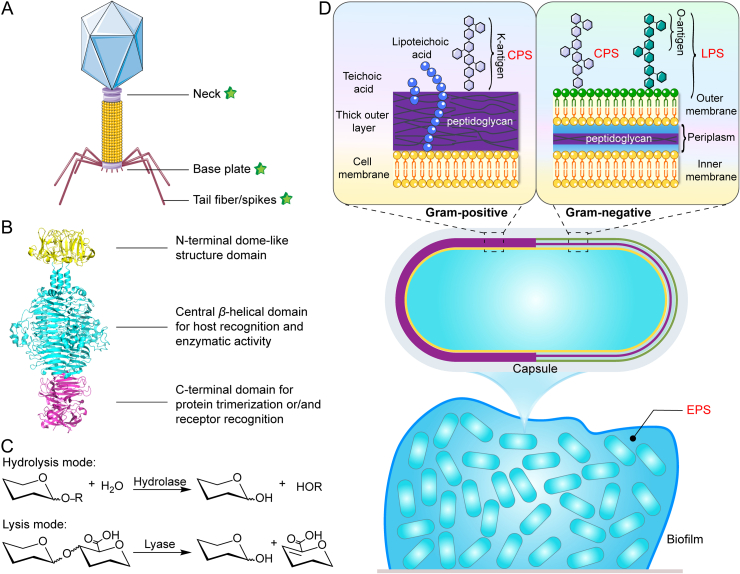
Figure 2.
Structure, mechanisms of action, and substrate specificity of depolymerases. (A) Distribution of depolymerase through the structure of bacteriophages. (B) The modular structure of the model tail spike (PDB ID 2XC1) of phage P22, consists of the N-terminal domain (yellow), central domain (cyan), and C-terminal domain (magentas). (C) General hydrolysis and lysis reaction mechanisms for depolymerases catalyzing polysaccharide degradation. (D) Varied targets of depolymerases in the architecture of bacteria and biofilms, including capsular polysaccharides (CPS), exopolysaccharides (EPS), and lipopolysaccharides (LPS).