ABSTRACT
Sphingomonadaceae are common membrane colonizers and biofilm formers. As part of our studies on long-term genetic changes in drinking water biofilm species, we report the draft genome sequence of Sphingomonas strain Sph5, isolated from a tap water filtration membrane. The isolate was determined as Sphingomonas paucimobilis through whole genome sequencing and de novo assembly.
KEYWORDS: draft genome, WGS, water, Sphingomonas
ANNOUNCEMENT
Widely used for drinking water, membrane filtration systems face biofouling due to microbe accumulation and biofilm formation. Studies reveal Sphingomonadaceae as common initial colonizers, persisting dominantly during biofilm growth (1 – 4). Here, we report the genome sequence of a prevalent isolate.
Sphingomonas spp. Sph5 was isolated from a biofilm on a Nadir MP005 microfiltration membrane used for drinking water biofouling studies (1, 5). Sph5 was previously reported to be phylogenetically closest to Sphingomonas sanguinis strains BAB-7166 (99%) and NBRC 13937 (99%) based on 16S rRNA sequencing and cultivation methods (1). However, the whole genome sequence (WGS) was not reported. WGS analysis is critical to uncover strain-specific traits associated with membrane biofouling. Here, we report the WGS of Sphingomonas spp. Sph5 using Illumina short-read sequencing and de novo assembly methods.
We received the Sphingomonas spp. Sph5 water isolate from Wetsus, Netherlands. Single isolated colonies were obtained on Reasoner’s 2A agar (R2A, Teknova R0005; Difco 214530) and cultured in R2A broth at room temperature. Genomic DNA was extracted using DNeasy UltraClean microbial kit (Qiagen 12224) following the company-provided protocol.
Sequencing libraries were generated using Kapa’s Hyperplus kit (KK8514) and IDT adapters (#00989130v2). Quality was verified using an Agilent Tapestation and qPCR (NEBNext Library Quant Kit, E7630L) on Thermo Fisher’s Quantstudio5.
Nineteen million 2 × 150 bp reads were obtained on a NovaSeq (Illumina) at Anschutz Medical Campus Genomics and Microarray Core, with an average Phred score of 33–36 calculated by FastQC [v0.11.9 (6)].
Assembly was performed with SPADES, default isolate settings [v3.15.2 (7)]. Contigs were aligned to NCBI databases with DIAMOND [v2.0.13 (8)]. Completeness was estimated using BUSCO [v5.2.2 (9)] proteobacteria marker genes. Of the 219 genes, there were 214 (97.7%) complete and single copy, 2 (0.9%) duplicated, 1 (0.5%) fragmented, and 2 (0.9%) missing.
For species identification, average nucleotide identity (ANI) was calculated between all complete Sphingomonas genomes and Sph5 with PYANI [v0.2.11 (10)] and MUMMER [v4.0.0 (11)]. ANI scores between Sph5 and S. paucimobilis ranged from 99.77%–99.95%, while ANI between Sph5 and S. sanguinis was 87.74%.
Annotations were predicted with Prokka [v1.14.6 (12)], given S. sanguinis and S. paucimobilis references (Table 1), and used for orthogroup-based phylogeny construction with Orthofinder [v2.5.4 (13)], MAFFT [v7.490 (14)], and FastTree [v.2.1.10 (15)].
TABLE 1.
Summary of the draft whole genome sequences of Sphingomonas paucimobilis Sph5 from the tap water filtration membrane
| Variable | Data |
|---|---|
| Genus and species | Sphingomonas paucimobilis |
| Strain | Sph5 |
| NCBI accession no. | JAMRJL000000000 |
| Country | The Netherlands |
| Source | Biofilm on Nadir MP005 microfiltration membrane |
| Feed water source | Tap water from the city of Leeuwarden |
| Genome size (bp) | 4680620 |
| N50 (bp) | 4016723 |
| Scaffold reference | ASM1602843v1 |
| Scaffolds ≥50,000 bp | 29, 4 |
| Mean coverage | 1,089× |
| G + C content (%) | 65.30 |
| No. of annotated CDS | 3,342 |
| No. of raw reads | 19,032,602 |
| No. of reads used for assembly | 19,032,602 |
| No. of coding sequences | 4,321 |
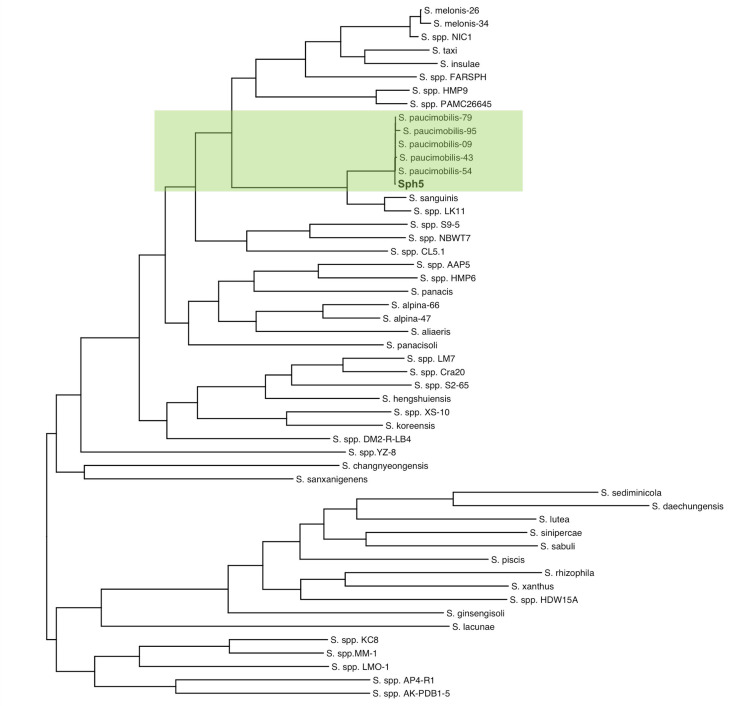
The WGS phylogeny (Fig. 1) confirms that Sph5 is closest to S. paucimobilis while related to S. sanguinis. As a result, S. paucimobilis reference genomes were used for RagTag [v2.1.0 (16)] scaffolding of Sph5 contigs. ASM1602843v1 was the best scaffold (93.6% total length in three longest contigs of resulting assembly). Realignment of raw reads to ASM1602843v1-scaffolded Sph5 assembly with BWA [v0.7.17 (17)], SamTools [v1.12 (18)], and Picard [v2.25.0 (19)], evaluated by Mosdepth [v0.3.3 (20)], supports this (99.48% reads aligned, 1,089× coverage).
Fig 1.
Orthofinder-generated rooted tree of 50 complete Sphingomonas genomes with the Sph5 strain. Branch lengths represent evolutionary distance. We used the multiple-sequence alignment option with MAFFT and used FastTree to infer the trees. The mathematical parameters used are built in to the OrthoFinder tool, and we did not alter them from the default.
Re-annotation with Prokka predicts 4,321 CDS, 3 rRNAs, 54 tRNAs, 1 tmRNA, and 1 repeat region. Of the CDS, 3,342 could be functionally annotated.
ACKNOWLEDGMENTS
This research was funded by NASA Space Biology, grant number 80NSSC19K1597, and NASA BAC, grant number HS5HWWK1AAU5.
Contributor Information
Jiseon Yang, Email: jyang41@asu.edu.
Julia A. Maresca, University of Delaware College of Engineering, Newark, Delaware, USA
DATA AVAILABILITY
The assembly and raw data are deposited at NCBI through the SRA genome submission portal under the accession numbers JAMRJL000000000 and SRR25337554. The version deposited in this paper is the first version.
REFERENCES
- 1. de Vries HJ, Beyer F, Jarzembowska M, Lipińska J, van den Brink P, Zwijnenburg A, Timmers PHA, Stams AJM, Plugge CM. 2019. Isolation and characterization of Sphingomonadaceae from fouled membranes. NPJ Biofilms Microbiomes 5:6. doi: 10.1038/s41522-018-0074-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koskinen R, Ali-Vehmas T, Kämpfer P, Laurikkala M, Tsitko I, Kostyal E, Atroshi F, Salkinoja-Salonen M. 2000. Characterization of Sphingomonas isolates from Finnish and Swedish drinking water distribution systems. J Appl Microbiol 89:687–696. doi: 10.1046/j.1365-2672.2000.01167.x [DOI] [PubMed] [Google Scholar]
- 3. Bereschenko LA, Stams AJM, Euverink GJW, van Loosdrecht MCM. 2010. Biofilm formation on reverse osmosis membranes is initiated and dominated by Sphingomonas spp. Appl Environ Microbiol 76:2623–2632. doi: 10.1128/AEM.01998-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gauthier V, Redercher S, Block JC. 1999. Chlorine inactivation of Sphingomonas cells attached to goethite particles in drinking water. Appl Environ Microbiol 65:355–357. doi: 10.1128/AEM.65.1.355-357.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dreszer C, Wexler AD, Drusová S, Overdijk T, Zwijnenburg A, Flemming H-C, Kruithof JC, Vrouwenvelder JS. 2014. In-situ Biofilm characterization in membrane systems using optical coherence tomography: formation, structure, detachment and impact of flux change. Water Res 67:243–254. doi: 10.1016/j.watres.2014.09.006 [DOI] [PubMed] [Google Scholar]
- 6. Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data
- 7. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- 9. Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. doi: 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- 10. Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. 2016. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods 8:12–24. doi: 10.1039/C5AY02550H [DOI] [Google Scholar]
- 11. Marçais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A. 2018. MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol 14:e1005944. doi: 10.1371/journal.pcbi.1005944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 13. Emms DM, Kelly S. 2019. Orthofinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20:238. doi: 10.1186/s13059-019-1832-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. doi: 10.1093/molbev/msp077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alonge M, Soyk S, Ramakrishnan S, Wang X, Goodwin S, Sedlazeck FJ, Lippman ZB, Schatz MC. 2019. RaGOO: fast and accurate reference-guided scaffolding of draft genomes. Genome Biol 20:224. doi: 10.1186/s13059-019-1829-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, Li H. 2021. Twelve years of SAMtools and BCFtools. Gigascience 10:giab008. doi: 10.1093/gigascience/giab008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. “Picard Toolkit” [Internet] . 2019. Broad Institute, Github repository. Available from: http://broadinstitute.github.io/picard
- 20. Pedersen BS, Quinlan AR. 2018. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics 34:867–868. doi: 10.1093/bioinformatics/btx699 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The assembly and raw data are deposited at NCBI through the SRA genome submission portal under the accession numbers JAMRJL000000000 and SRR25337554. The version deposited in this paper is the first version.