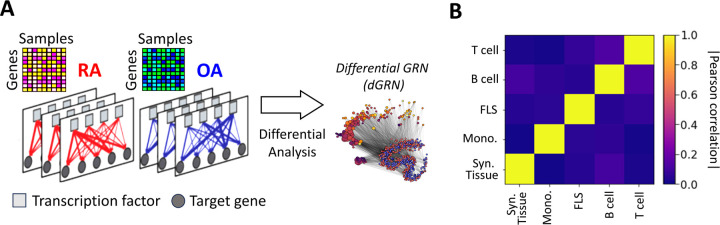
Figure 2:
(A) Networks were inferred from the gene expression profiles of RA and OA biopsies in different cell types. Network edges are endowed with weights representing the probability of regulatory interactions between a transcription factor (TF) and a target gene (TG). The analysis of differences in edge weights between RA and OA facilitated the construction of a differential GRN (dGRN) for each cell type. The dGRN was used to compute a regulatory score for each TF in each cell type. (B) Heatmap of the Pearson correlation between the TF regulatory scores in each tissue type, i.e. synovial tissue, monocyte, FLS, B cell, and T cell).