FIGURE 6.
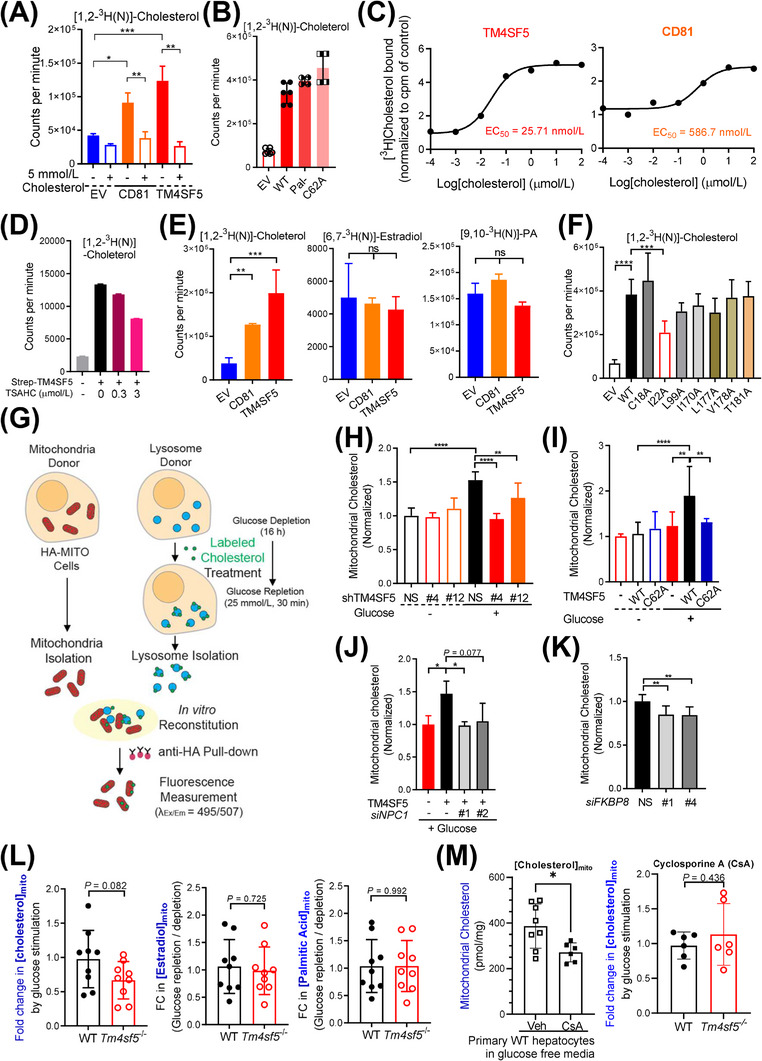
TM4SF5‐enriched MLCSs export lysosomal cholesterol to mitochondria. (A‐D) Huh7 cells (A, C, and D) or SNU449 cells (B) transfected with the indicated expression plasmids were harvested, and the whole‐cell lysates were pulled down with streptavidin beads. The beads were washed and incubated with 5 μmol/L radiolabeled cholesterol with 5 mmol/L cold cholesterol (A) or without (B), different concentrations of radiolabeled cholesterol alone (C), or with TSAHC treatment (D), before CPM measurements. (E) Immunopurified CD81‐Strep or TM4SF5‐Strep prepared as in (A) was incubated with [1,2‐3H(N)]‐cholesterol, [6,7‐3H(N)]‐estradiol, or [9,10‐3H(N)]‐palmitic acid. Bound 3H‐lipid was measured in a scintillation counter. (F) SNU449 cells transfected with indicated expression plasmids were processed for [1,2‐3H(N)]‐cholesterol binding assay as in (A). (G) Schematic presentation of the In vitro mitochondria‐lysosome contact assay. (H‐K) Huh7 stably expressing HA‐OMP25 were processed for rapid mitochondria isolation by anti‐HA pulldown. Lysosomes were isolated by ultracentrifugation from another set of stable Huh7 cells with or without TM4SF5 suppression (H), SNU449 cells transfected with Strep‐EV or TM4SF5 WT or C62A (I), SNU449 cells transfected with TM4SF5‐FLAG and/or siNPC1 (against NPC1 sequences #1 and #2, J) were glucose depleted (‐) and then replete (+) with 10 μmol/L of TopFluor cholesterol treatment (2 h). After the isolated organelles were mixed for 30 min at 37°C, anti‐HA antibody beads were used to isolate the mitochondria for fluorescence measurements. Huh7 cells stably expressing HA‐OMP25 were treated with siRNA targeting FKBP8 (against #1 and #4 sequences) and processed for mitochondria isolation and In vitro mitochondria‐lysosome contact assay (K). (L‐M) Primary hepatocytes from WT and Tm4sf5 −/‐ KO mice infected with HA‐OMP25 lentivirus were glucose‐depleted for 16 h. Thirty minutes before cell harvest, 25 mmol/L glucose was replete (L). In case, the primary cells were treated with cyclosporine A 10 μmol/L for 24 h to impair lysosomes (M). The cells were then suspended in PBS and mildly homogenized before pulldown using anti‐HA‐magnetic beads, methanol extraction, and GC/MS‐based cholesterol, estradiol, or palmitate measurement. Fold changes were calculated from the values of repletion samples divided by the values from depletion samples. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, ns = non‐significant. Data were represented as mean ± SD. Data represent three independent experiments. See also Supplementary Figure S8. CPM, counter per minute; FKBP8, FK506‐binding protein 8; HA‐OMP25, outer membrane protein 25 for tagging mitochondria with HA epitopes; KO, knockout; MLCS, mitochondria‐lysosome contact site; SD, standard deviation; TM4SF5, transmembrane 4 L six family member 5; TSAHC,4'‐(p‐toluenesulfonylamido)‐4‐hydroxychalcone; WT, wild‐type.
