Figure 2. Heterochromatin domains and synaptic gene silencing on autosomes in FXS patient-derived iPSC-NPCs and brain tissue.
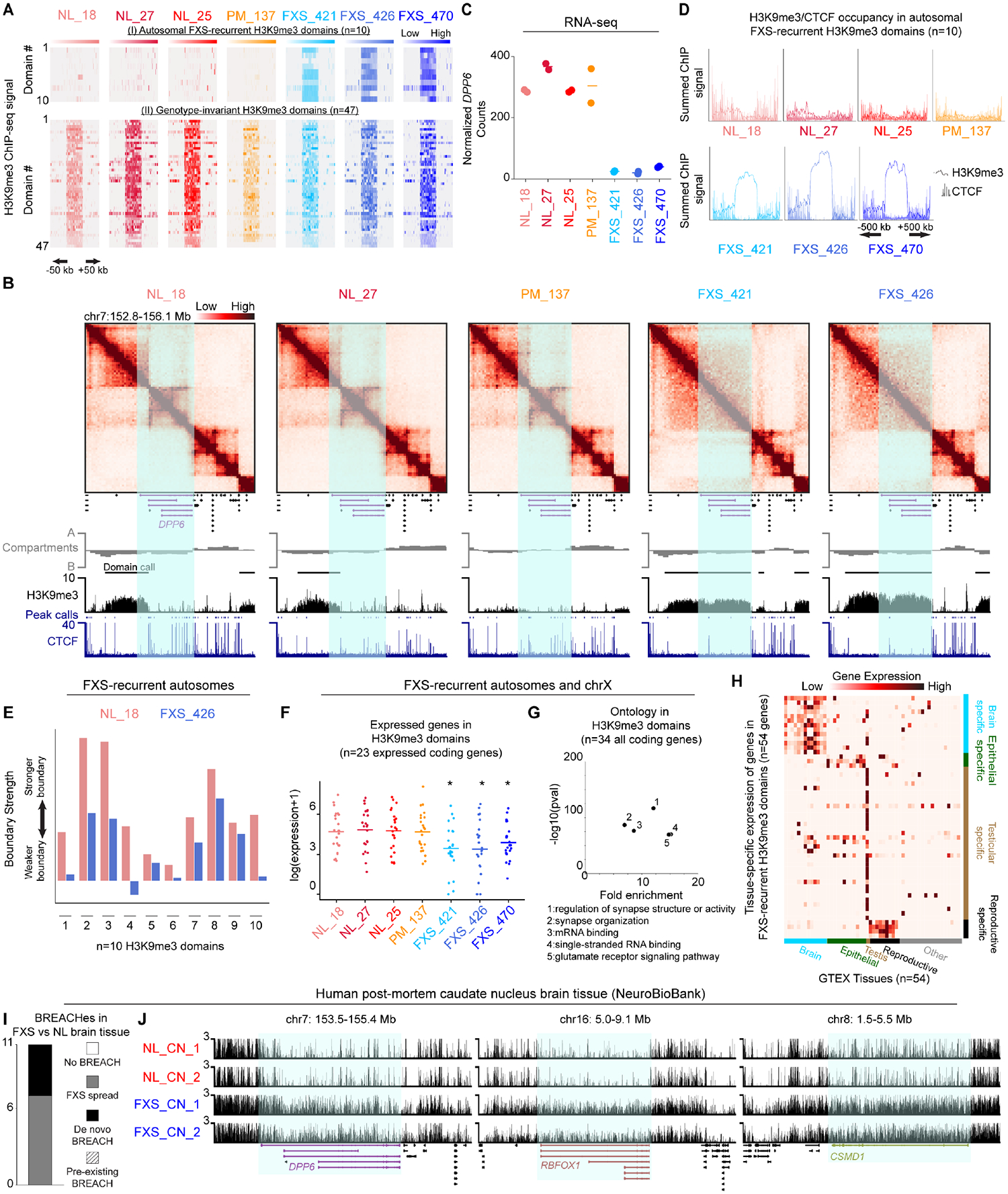
(A) Two classes of autosomal H3K9me3 domains (i) FXS-recurrent: consistently gained in all three FXS iPSC-NPCs and not in NL/PM iPSC-NPCs or (ii) Genotype-invariant: present in NL/PM/FXS iPSC-NPCs. (B) Hi-C and ChIP-seq for a 3.5 Mb region around a H3K9me3 domain encompassing DPP6. (C) DPP6 mRNA levels via RNA-seq. Horizontal lines, mean n=2 biological replicates. (D) Average H3K9me3 and CTCF ChIP-seq signal across autosomal FXS-recurrent H3K9me3 domains. (E) Boundary strength in NL_18 and FXS_426 iPSC-NPCs for one TAD boundary per autosomal FXS-recurrent H3K9me3 domain. (F) mRNA levels via RNA-seq for N=25 expressed protein-coding genes in autosomal and chrX FXS-recurrent H3K9me3 domains. Each point, mean per gene n=2 biological replicates. P-values, one-tailed MWU, where * P-value <0.05 versus NL_18. (G) Gene ontology for all N=36 protein-coding genes in autosomal and chrX FXS-recurrent H3K9me3 domains. (H) Expression of N=54 coding/noncoding genes in FXS-recurrent H3K9me3 domains across GTEX tissues. (I) Number of autosomal H3K9me3 domains arising in FXS patient-derived brain tissue compared to sex- and age-matched normal-length control tissue. (J) H3K9me3 CUT&RUN in brain tissue from N=2 FXS patients with sex- and age-matched N=2 normal-length individuals at DPP6, RBFOX1, and CSMD1.
