Figure 3. Engineering the mutation-length FMR1 CGG STR to premutation-length attenuates a subset of H3K9me3 domains and de-represses gene expression.
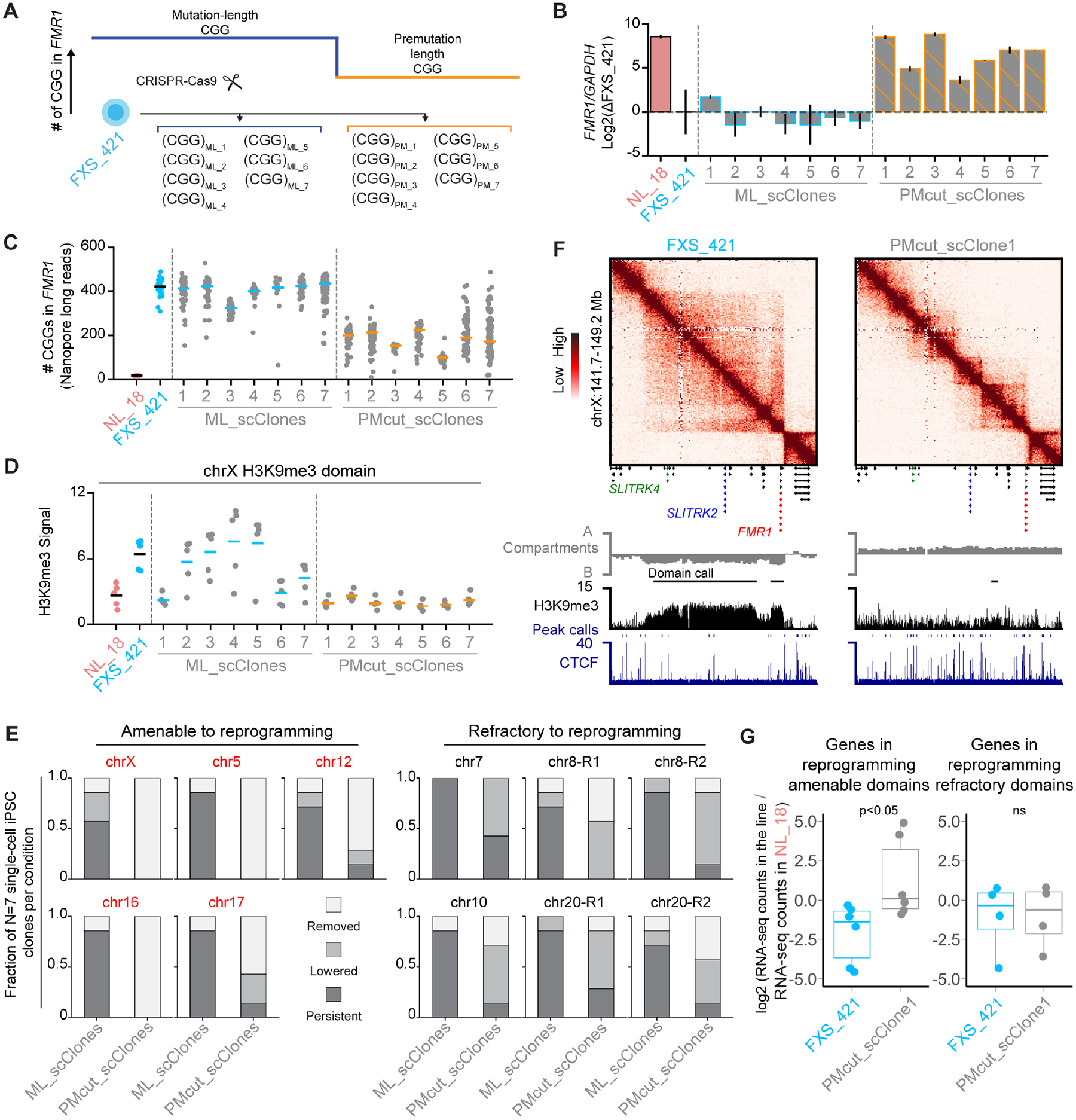
(A) Schematic of N=7 mutation-length and premutation-length single-cell-derived CGG CRISPR cut-back iPSC clones generated from the FXS_421 parent iPSC line. (B) FMR1 mRNA levels normalized to GAPDH and shown relative to FXS_421 using qRT-PCR. Error bars, standard deviation n=2 biological replicates. (C) Number of CGG triplets in the FMR1 5’UTR computed from Nanopore long-reads. (D) Average input normalized H3K9me3 signal for the chrX FXS-recurrent H3K9me3 domain. Dots represent equal sized bins (N=5) across the domain. (E) FXS-recurrent H3K9me3 domains amenable (red) and refractory (black) to reprogramming. For each domain, we measured the fraction of iPSC clones with persistent, lowered, or removed H3K9me3 signal for all mutation-length (N=7) and premutation-length (N=7) clones. (F) Hi-C and ChIP-seq for a 5 Mb region around FMR1 in FXS_421 and PMcut_scClone1 iPSCs. (G) Log2 fold change of gene expression in FXS_421 vs. PMcut_scClone1 with respect to NL_18. Each dot, one gene. P-values, one-tailed MWU.
