Figure 4. Autosomal heterochromatin domains spatially connect with FMR1 via inter-chromosomal interactions in FXS.
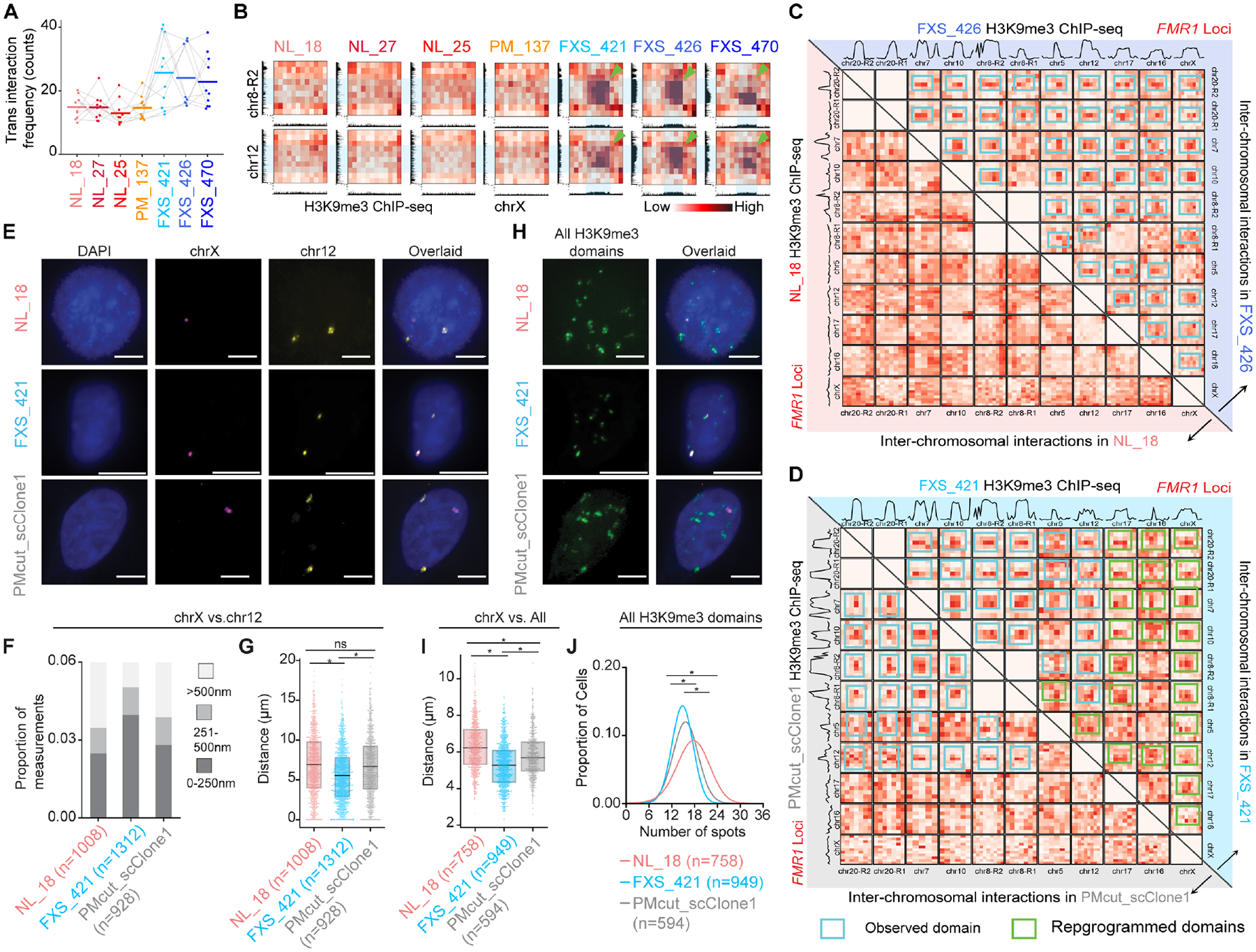
(A) Trans interactions between each of the N=10 FXS-recurrent H3K9me3 domain on autosomes and FMR1 on chrX. (B) Hi-C inter-chromosomal interaction heatmaps binned at 1 Mb resolution. Green arrows, trans interactions. (C-D) Hi-C inter-chromosomal interactions among FXS-recurrent H3K9me3 domains (C) FXS_426 (upper triangle) versus NL_18 (lower triangle) iPSC-NPCs and (D) FXS_421 (upper triangle) versus PMcut_scClone1 (lower triangle) iPSCs. H3K9me3 ChIP-seq signal plotted above Hi-C heatmaps. Blue boxes, FXS-gained trans interactions. Green boxes, attenuated trans interactions after premutation-length cutback. (E+H) DNA FISH images for the H3K9me3 domain on chrX interacting with (E) the chr12 domain or (H) all domains in NL_18, FXS_421, and PMcut_scClone1 iPSC nuclei. Scale bars, 10 μm. (F-G) Distances between chrX and chr12 H3K9me3 domains in iPSCs, including (F) proportion of measurements stratified by distance and (G) measurements directly compared with a two-tailed MWU, where * P-value <1E-6. (I) Average distance per cell between the chrX and all other FXS-recurrent H3K9me3 domains. (J) Kernel density estimation of the number of foci per nucleus. (I-J) Two-tailed MWU, where * P-value <1E-12.
