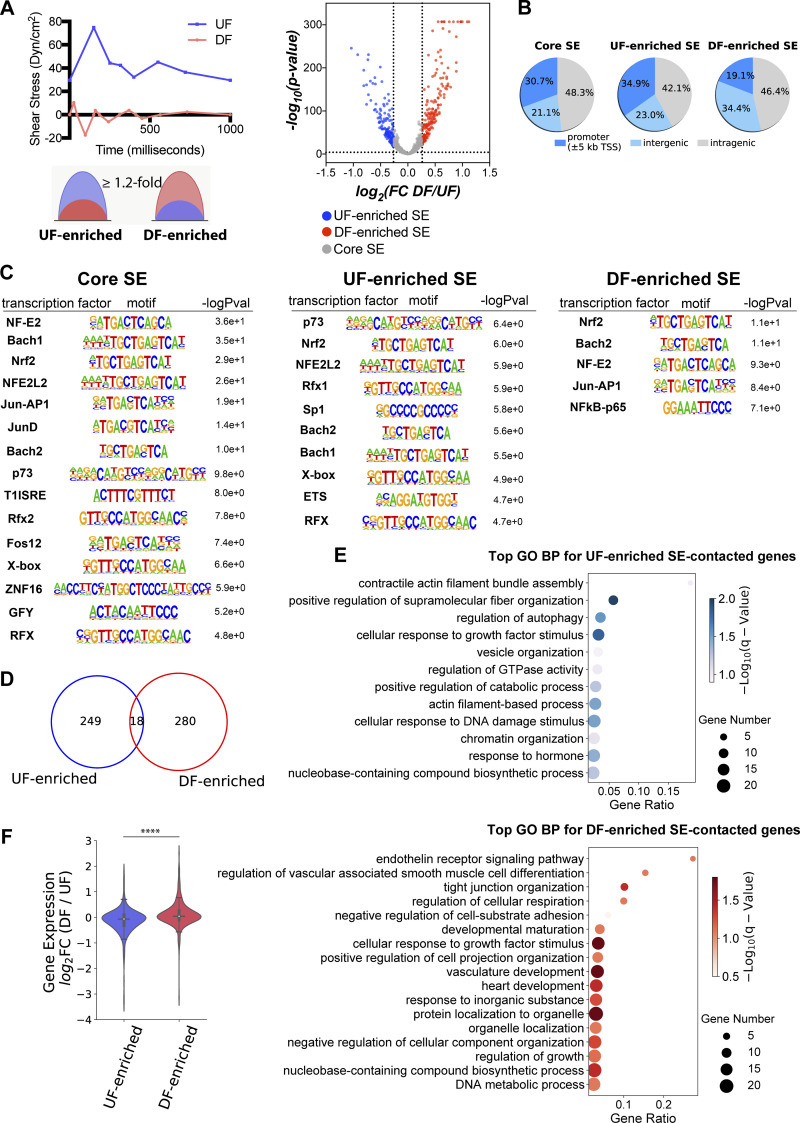
Figure 2.
Flow-sensitive endothelial super-enhancers preferentially contact the promoters of flow-sensitive genes to regulate their transcription. (A) Classification of UF-enriched super-enhancers and DF-enriched super-enhancers in HAECs. Top left: Athero-relevant shear stress was generated by a dynamic flow device and applied to cultured HAECs. Athero-protective UF (blue) and athero-susceptible DF (red) represent hemodynamics in the human distal internal carotid artery and carotid sinus, respectively. Bottom left: Schematic plots depicting UF-enriched or DF-enriched super-enhancers, of which the fold change of H3K27ac signal responding to flows is ≥1.2. Right: volcano plot demonstrating the flow-induced H3K27ac signal change at each super-enhancer locus. UF-enriched super-enhancers (blue dots) are determined by log2(fold change, DF/UF) less-than or equal to −0.263 and −log10(P value) ≥4; DF-enriched super-enhancers (red dots) are determined by log2(fold change, DF/UF) ≥0.263 and −log10(P value) ≥4; the complementary sets are defined as core super-enhancers (gray dots) with |log2(fold change, DF/UF)| <0.263. (B) Pie charts of genomic distributions of core, UF-enriched, and DF-enriched endothelial super-enhancers. (C) Enriched transcription factor binding motifs in core, UF-enriched, and DF-enriched endothelial super-enhancers. (D) The Venn diagram demonstrating the number of genes uniquely contacted by either UF-enriched or DF-enriched super-enhancers, and genes contacted by both types of super-enhancers. (E) Gene ontology analyses revealed biological processes of genes exclusively contacted by either UF-enriched super-enhancers (top) or DF-enriched super-enhancers (bottom). (F) Flow-induced endothelial super-enhancers positively correlated with flow-induced transcription. RNA-seq results demonstrate that the majority of genes contacted by DF-enriched super-enhancers have increased transcription levels in HAECs under DF when compared to cells under UF (median log2FC > 0, DF/UF), whereas the majority of genes contacted by UF-enriched super-enhancers are transcriptionally upregulated by UF (median log2FC < 0, DF/UF). The average transcriptional fold change is significantly different between genes contacted by two clusters of flow-sensitive super-enhancers. Quartiles were represented by black lines. ****P value ≤0.0001 was determined by a two-sided Student’s t test.