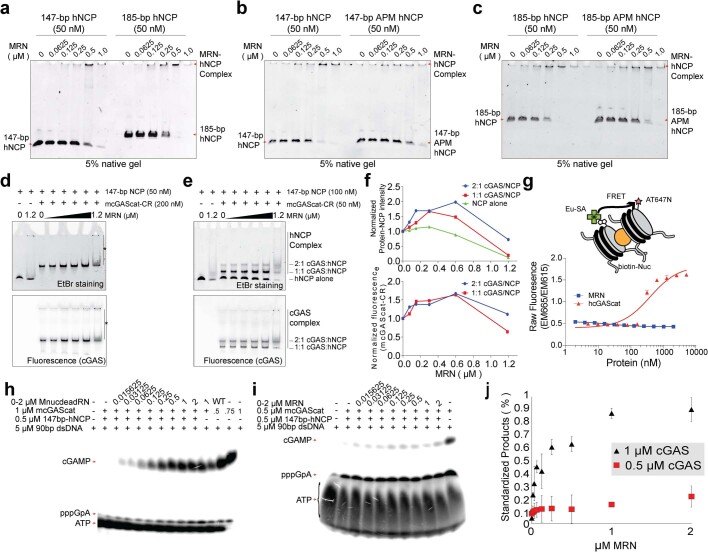
Extended Data Fig. 7. Biochemical analysis of purified MRN, Nucleosome Core Particles (NCP), and cGAS.
a, b, c, d, Electrophoretic mobility shift assays (EMSA) a, Native gel of nucleosomes containing 147 or 185 bp DNA in presence of increasing concentrations of MRN complex, stained with ethidium bromide. b, Native gel of 147 bp nucleosomes containing either wild type or acidic patch mutant (APM) histones in the presence of increasing concentrations of MRN complex, stained with ethidium bromide. c, Native gel as in panel (b) using 185 bp nucleosomes with 20 bp symmetric linker on each side of the nucleosome core. d and e, Native gel of 50 nM (d) or 100 nM (e) 147-bp hNCPs without or with 200 nM (d) or 50 nM (e) carboxyrhodamine-labelled mouse cGAS catalytic domain (mcGAScat-CR) and 0-1.2 μM MRN. The upper gel shows ethidium bromide staining of DNA, and the lower gel shows fluorescence signal from mcGAScat-CR. hNCP alone as well as 1:1 and 2:1 cGAS:hNCP complexes are indicated. (The results from the tests (a, b, c, d, e) demonstrate consistent findings across three independent replicates. f, Quantification of bands from the corresponding gels (e). g, TR-FRET assay for detection of nucleosome stacking mediated by human cGAS catalytic domain (hcGAScat) or MRN complex. EU-SA, LANCE Eu-W1024 Streptavidin; AT647N, Atto647N; biotin-NUC, H2BK125C-biotin 147-bp nucleosomes. Data are mean ± s.d. h, i, Substrate, intermediate, and product are ATP, pppGpA, and cGAMP, respectively. h, 1 μM cGAS: 5 μM dsDNA, 0.5 μM hNCP, 0–2 μM MnucdeadRN (containing Mre11H129N nuclease deficient protein), demonstrating consistent findings across three independent replicates. i, 0.5 μM cGAS: 5 μM dsDNA, 0.5 μM hNCP, 0–2 μM MRN demonstrating consistent findings across two independent replicates. j, Quantification of standardized percentage of cGAMP product across three biological replicates demonstrates greater cGAS activity in the presence of increasing concentrations of MRN, but only when the cGAS:hNCP ratio is 2:1 versus 1:1. Shown are the mean value with error bars representing the standard deviation.