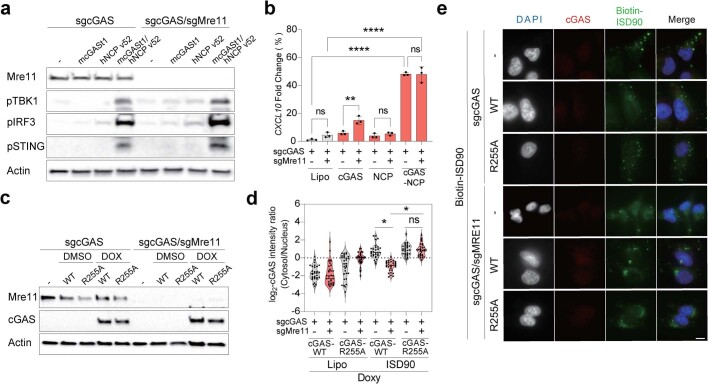
Extended Data Fig. 9. Mre11 is required to release cGAS from nucleosome tethering.
a, b, Human cGAS knock-out MDA-MB231 cell lines were made with CRISPR technology using multiguided sgRNA (Synthego). Then, MDA-MB-231 sgcGAS stable cell lines were transfected with mcGASt1, hNCP v52, or mcGASt1/hNCP v52. a, After 2 h, cells were harvested for Western blot analysis to confirm human Mre11, pTBK, pSTING, and Actin, demonstrating consistent findings across two independent replicates. b, Six hours after transfection, qRT-PCR was performed to normalize gene expression levels for interferon-stimulated genes CXCL10, with mRNA levels normalized to β-actin mRNA levels. n = 3 independent experiments; 3 samples for each cell lines. sgcGAS + Lipo vs sgcGAS + cGAS-NCP; p < 0.0001, sgcGAS/sgMRE11 + Lipo vs sgcGAS/sgMRE11 + cGAS-NCP; p < 0.0001, sgcGAS + cGAS vs sgcGAS/sgMRE11+ cGAS; p = 0.0067. c, d, PiggyBac Transposon system was used to deliver PB-cGAS-WT or PB-cGAS-R255A (an AP site binding mutant) to each cell line, followed by selection with puromycin for two days. After selection, cells were incubated with doxycycline for 24 h, and then transfected with ISD90. c, Western blot analysis of human Mre11, cGAS and Actin 24 h after doxycycline treatment, demonstrating consistent findings across two independent replicates. d, The cGAS localization ratio (Nucleus/cytosol) 2 h after Biotin-ISD90 transfection. n = 2 independent experiments; 31 cells for each cell lines. sgcGAS + cGAS-WT + ISD90 vs sgcGAS/sgMRE11 + cGAS-WT + ISD90; p = 0.0394, sgcGAS/sgMRE11 + cGAS-WT + ISD90 vs ssgcGAS/sgMRE11 + cGAS-R255A + ISD90; p = 0.022. e, Representative images showing the localization of cGAS and Biotin-ISD90 were obtained 2 h after transfection with Biotin-ISD90, demonstrating consistent findings across two independent replicates. Scale bar, 10 μm. Data are mean ± SEM. Unless otherwise specified, statistical analyses were determined by one-way ANOVA followed by Sidak’s multiple comparison post-test using a two-tailed test: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. n.s., not significant.