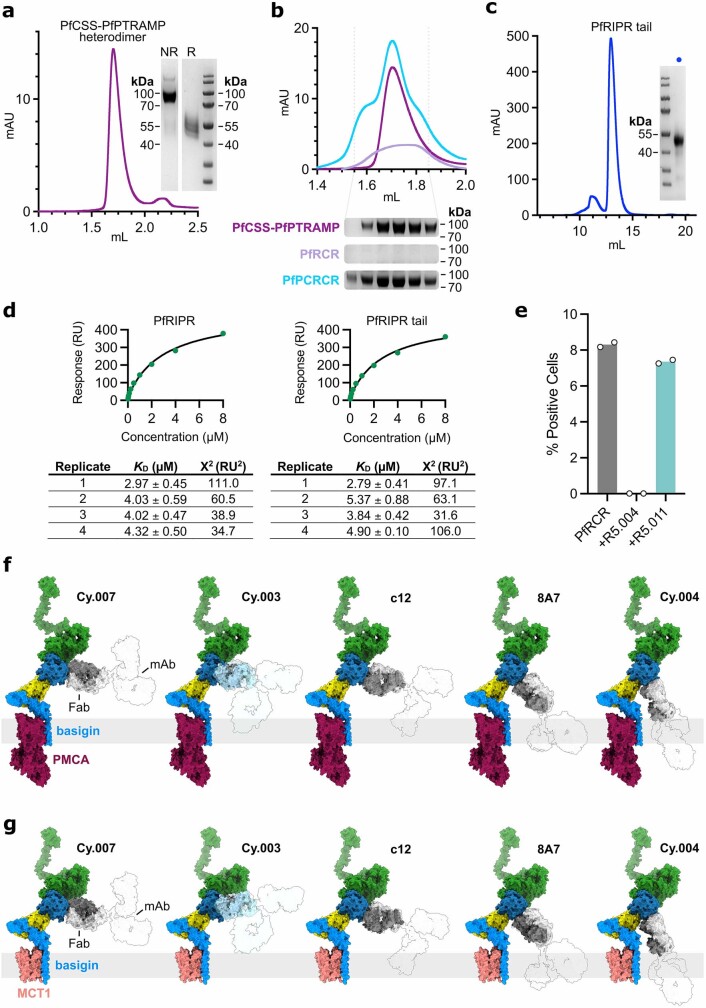
Extended Data Fig. 8. PfRIPR tail interacts with the PfCSS-PfPTRAMP complex, and the mapping of anti-PfCyRPA antibodies.
a, Gel filtration trace of the PfCSS-PfPTRAMP heterodimer with an SDS-PAGE gel inset right. SDS-PAGE fractions are shown for non-reduced (NR) and reduced (R) samples of the disulphide-linked heterodimer. Representative of three independent experiments. b, Gel filtration traces for PfCSS-PTRAMP complex (dark purple), PfRCR (light purple) and the mixture of the two complexes (blue). Underneath non-reducing SDS-PAGE gels show the corresponding fractions for the location of PfCSS-PfPTRAMP from the three experiments. Traces and gels are representative of two independent analyses. c, Gel filtration trace of recombinantly expressed PfRIPR tail with reducing SDS-PAGE gel of isolated PfRIPR tail shown inset right, representative of three independent samples. d, Representative steady state fit analysis of PfCSS-PfPTRAMP complex binding to full-length PfRIPR and PfRIPR tail by SPR. Binding analysis was completed four times. Inset below are the estimated binding affinity (KD) and the Chi2 fit for each repeat. e, FACS-based analysis of the binding of PfRCR complex to human erythrocytes in the presence of PfRH5-targeting antibodies R5.004 and R5.011. In each case, PfRCR was detected using an anti-C-tag nanobody targeting the affinity tag of the complex. Binding of the PfRCR complex is unaffected by the presence of R5.011. N = 2, individual data points, the mean +/- SD are shown. f, Composite model showing structural alignment of anti-PfCyRPA antibodies on to the PfRCR complex when bound to PMCA-bound basigin on the erythrocyte surface, with the rest of the antibodies modelled as faint silhouettes. Generated through alignment of the PfRCR-Cy.003 complex, Fab-PfCyRPA crystal structures (PDB IDs: 7PHV, 5EZO, 5TIH and 7PHW)5,19,20, an intact IgG crystal structure (PDB ID: 1IGT), and PMCA-bound basigin (PDB ID: 6A6934). g, As in (f) except when PfRCR is bound to MCT1-bound basigin (PDB ID: 7CKR33).